3ABI
 
 | | Crystal Structure of L-Lysine Dehydrogenase from Hyperthermophilic Archaeon Pyrococcus horikoshii | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative uncharacterized protein PH1688, SULFATE ION | | Authors: | Yoneda, K, Sakuraba, H, Fukuda, J, Ohshima, T. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | First crystal structure of L-lysine 6-dehydrogenase as an NAD-dependent amine dehydrogenase.
J.Biol.Chem., 285, 2010
|
|
4PJ2
 
 | | Crystal structure of Aeromonas hydrophila PliI in complex with Meretrix lusoria lysozyme | | Descriptor: | GLYCEROL, Lysozyme, MAGNESIUM ION, ... | | Authors: | Leysen, S, Van Herreweghe, J.M, Yoneda, K, Ogata, M, Usui, T, Michiels, C.W, Araki, T, Strelkov, S.V. | | Deposit date: | 2014-05-10 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The structure of the proteinaceous inhibitor PliI from Aeromonas hydrophila in complex with its target lysozyme.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7XY8
 
 | | Crystal structure of antibody Fab fragment in complex with CD147(EMMPIRIN) | | Descriptor: | Isoform 2 of Basigin, heavy chain, light chain | | Authors: | Nakamura, K, Amano, M, Yoneda, K, Suzuki, M, Fukuchi, K. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Antibody Exerts Antitumor Effect through Downregulation of CD147 and Activation of Multiple Stress Signals.
J Oncol, 2022, 2022
|
|
4XB2
 
 | | Hyperthermophilic archaeal homoserine dehydrogenase mutant in complex with NADPH | | Descriptor: | 319aa long hypothetical homoserine dehydrogenase, L-HOMOSERINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
4XB1
 
 | | Hyperthermophilic archaeal homoserine dehydrogenase in complex with NADPH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 319aa long hypothetical homoserine dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
3ICP
 
 | |
3KO8
 
 | | Crystal Structure of UDP-galactose 4-epimerase | | Descriptor: | NAD-dependent epimerase/dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Sakuraba, H, Kawai, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2009-11-13 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of UDP-galactose 4-epimerase from the hyperthermophilic archaeon Pyrobaculum calidifontis
Arch.Biochem.Biophys., 512, 2011
|
|
5FB3
 
 | | Structure of glycerophosphate dehydrogenase in complex with NADPH | | Descriptor: | Glycerol-1-phosphate dehydrogenase [NAD(P)+], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYROPHOSPHATE, ... | | Authors: | Sakuraba, H, Hayashi, J, Yamamoto, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Unique coenzyme binding mode of hyperthermophilic archaeal sn-glycerol-1-phosphate dehydrogenase from Pyrobaculum calidifontis
Proteins, 84, 2016
|
|
6K9Z
 
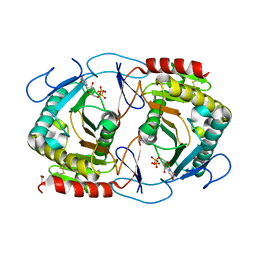 | | STRUCTURE OF URIDYLYLTRANSFERASE MUTANT | | Descriptor: | ACETATE ION, FE (III) ION, Galactose-1-phosphate uridylyltransferase, ... | | Authors: | Sakuraba, H, Ohshida, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-06-19 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Unique active site formation in a novel galactose 1-phosphate uridylyltransferase from the hyperthermophilic archaeon Pyrobaculum aerophilum.
Proteins, 88, 2020
|
|
6K5Z
 
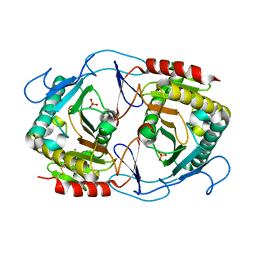 | | Structure of uridylyltransferase | | Descriptor: | FE (III) ION, Galactose-1-phosphate uridylyltransferase, PHOSPHATE ION, ... | | Authors: | Sakuraba, H, Ohshida, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-05-31 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Unique active site formation in a novel galactose 1-phosphate uridylyltransferase from the hyperthermophilic archaeon Pyrobaculum aerophilum.
Proteins, 88, 2020
|
|
5GZ6
 
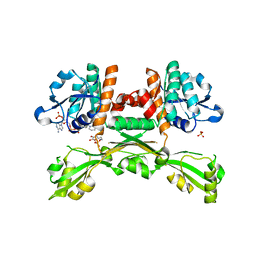 | | Structure of D-amino acid dehydrogenase in complex with NADPH and 2-keto-6-aminocapronic acid | | Descriptor: | 6-azanyl-2-oxidanylidene-hexanoic acid, ACETATE ION, Meso-diaminopimelate D-dehydrogenase, ... | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5GZ3
 
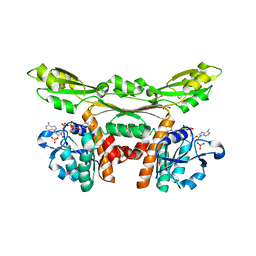 | | Structure of D-amino acid dehydrogenase in complex with NADP | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5GZ1
 
 | | Structure of substrate/cofactor-free D-amino acid dehydrogenase | | Descriptor: | Meso-diaminopimelate D-dehydrogenase | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
3WQO
 
 | | Crystal structure of D-tagatose 3-epimerase-like protein | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein MJ1311 | | Authors: | Uechi, K, Takata, G, Yoneda, K, Ohshima, T, Sakuraba, H. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of D-tagatose 3-epimerase-like protein from Methanocaldococcus jannaschii
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3VQR
 
 | | Structure of a dye-linked L-proline dehydrogenase mutant from the aerobic hyperthermophilic archaeon, Aeropyrum pernix | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sakuraba, H, Ohshima, T, Satomura, T, Yoneda, K. | | Deposit date: | 2012-03-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Novel Dye-linked L-Proline Dehydrogenase from Hyperthermophilic Archaeon Aeropyrum pernix
J.Biol.Chem., 287, 2012
|
|
3VTF
 
 | | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Sakuraba, H, Ohshima, T, Yoneda, K. | | Deposit date: | 2012-05-29 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6K3D
 
 | | Structure of multicopper oxidase mutant | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, Multicopper oxidase | | Authors: | Sakuraba, H, Ohshida, T, Satomura, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Activity enhancement of multicopper oxidase from a hyperthermophile via directed evolution, and its application as the element of a high performance biocathode.
J.Biotechnol., 325, 2021
|
|
3WID
 
 | | Structure of a glucose dehydrogenase T277F mutant in complex with NADP | | Descriptor: | Glucose 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WVX
 
 | |
3WIC
 
 | | Structure of a substrate/cofactor-unbound glucose dehydrogenase | | Descriptor: | Glucose 1-dehydrogenase, PENTAETHYLENE GLYCOL, S-1,2-PROPANEDIOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WYH
 
 | |
3WIE
 
 | | Structure of a glucose dehydrogenase T277F mutant in complex with D-glucose and NAADP | | Descriptor: | Glucose 1-dehydrogenase, ZINC ION, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(3-carboxypyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WVY
 
 | | Structure of D48A hen egg white lysozyme in complex with (GlcNAc)4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | Authors: | Kawaguchi, Y, Yoneda, K, Araki, T. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The role of Asp48 in the hydrogen bonding network involving Asp52 of hen egg white lysozyme
TO BE PUBLISHED
|
|
3A9H
 
 | | Crystal Structure of PQQ-dependent sugar dehydrogenase holo-form | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, Putative uncharacterized protein, ... | | Authors: | Sakuraba, H, Yokono, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2009-10-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic properties and crystal structure of quinoprotein aldose sugar dehydrogenase from hyperthermophilic archaeon Pyrobaculum aerophilum
Arch.Biochem.Biophys., 502, 2010
|
|
3A9G
 
 | | Crystal Structure of PQQ-dependent sugar dehydrogenase apo-form | | Descriptor: | CALCIUM ION, Putative uncharacterized protein, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Sakuraba, H, Yokono, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2009-10-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Catalytic properties and crystal structure of quinoprotein aldose sugar dehydrogenase from hyperthermophilic archaeon Pyrobaculum aerophilum
Arch.Biochem.Biophys., 502, 2010
|
|
