2RV3
 
 | | Solution structures of the DNA-binding domain (ZF15) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RUV
 
 | | Solution structures of the DNA-binding domain (ZF4) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RV5
 
 | | Solution structures of the DNA-binding domain (ZF8) of mouse immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RUU
 
 | | Solution structures of the DNA-binding domain (ZF3) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RUZ
 
 | | Solution structures of the DNA-binding domain (ZF11) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RR6
 
 | | Solution structure of the leucine rich repeat of human acidic leucine-rich nuclear phosphoprotein 32 family member B | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Authors: | Tochio, N, Umehara, T, Tsuda, K, Koshiba, S, Harada, T, Watanabe, S, Tanaka, A, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
J.Mol.Biol., 401, 2010
|
|
2RV7
 
 | | Solution structures of the DNA-binding domains (ZF3-ZF4-ZF5) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RRB
 
 | | Refinement of RNA binding domain in human Tra2 beta protein | | Descriptor: | cDNA FLJ40872 fis, clone TUTER2000283, highly similar to Homo sapiens transformer-2-beta (SFRS10) gene | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Sugano, S, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the dual RNA-recognition modes of human Tra2-beta RRM.
Nucleic Acids Res., 39, 2011
|
|
3JQK
 
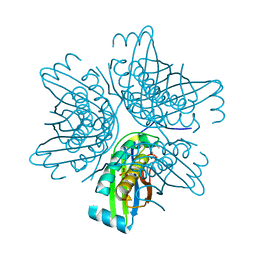 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 (H32 FORM) | | Descriptor: | ACETATE ION, Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3JQJ
 
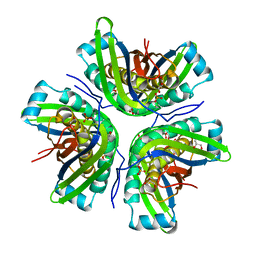 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | Descriptor: | GLYCEROL, Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION, ... | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
1G59
 
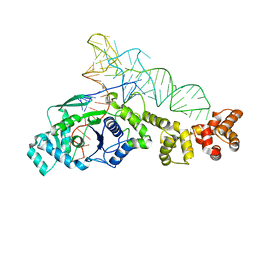 | | GLUTAMYL-TRNA SYNTHETASE COMPLEXED WITH TRNA(GLU). | | Descriptor: | GLUTAMYL-TRNA SYNTHETASE, TRNA(GLU) | | Authors: | Sekine, S, Nureki, O, Shimada, A, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-10-31 | | Release date: | 2001-09-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase.
Nat.Struct.Biol., 8, 2001
|
|
1GLN
 
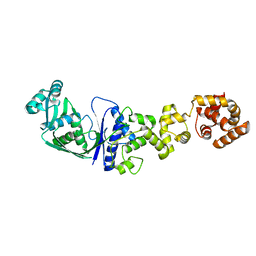 | | ARCHITECTURES OF CLASS-DEFINING AND SPECIFIC DOMAINS OF GLUTAMYL-TRNA SYNTHETASE | | Descriptor: | GLUTAMYL-TRNA SYNTHETASE | | Authors: | Nureki, O, Vassylyev, D.G, Katayanagi, K, Shimizu, T, Sekine, S, Kigawa, T, Miyazawa, T, Yokoyama, S, Morikawa, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1994-07-20 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architectures of class-defining and specific domains of glutamyl-tRNA synthetase.
Science, 267, 1995
|
|
2ROZ
 
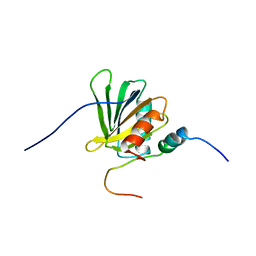 | | Structure of the C-terminal PID Domain of Fe65L1 Complexed with the Cytoplasmic Tail of APP Reveals a Novel Peptide Binding Mode | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 2, peptide from Amyloid beta A4 protein | | Authors: | Li, H, Koshiba, S, Tochio, N, Watanabe, S, Harada, T, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
3I7V
 
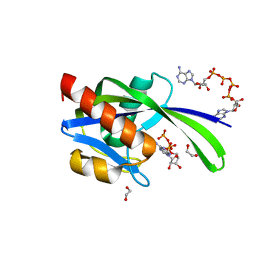 | | Crystal structure of AP4A hydrolase complexed with AP4A (ATP) (aq_158) from Aquifex aeolicus Vf5 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, AP4A hydrolase, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Nakagawa, N, Sekar, K, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Free and ATP-bound structures of Ap(4)A hydrolase from Aquifex aeolicus V5
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3I7U
 
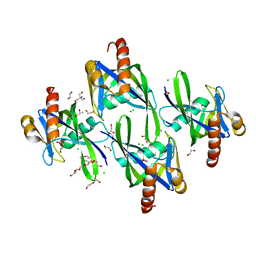 | | Crystal structure of AP4A hydrolase (aq_158) from Aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AP4A hydrolase, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Nakagawa, N, Sekar, K, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Free and ATP-bound structures of Ap(4)A hydrolase from Aquifex aeolicus V5
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3JQM
 
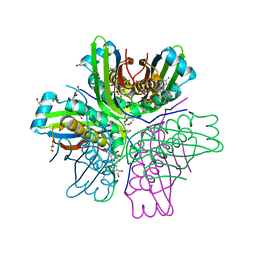 | | Binding of 5'-GTP to molybdenum cofactor biosynthesis protein MoaC from Thermus theromophilus HB8 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3HJN
 
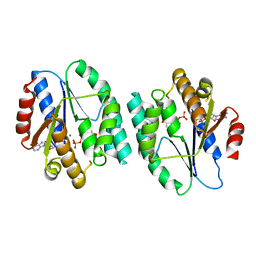 | | Crystal structure of thymidylate kinase in complex with dTDP and ADP from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Yoshikawa, S, Nakagawa, N, Shirouzu, M, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymidylate kinase in complex with dTDP and ADP from Thermotoga maritima
To be Published
|
|
2RU3
 
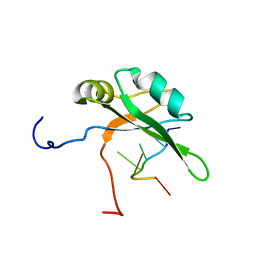 | | Solution structure of c.elegans SUP-12 RRM in complex with RNA | | Descriptor: | Protein SUP-12, isoform a, RNA (5'-R(*GP*UP*GP*UP*GP*C)-3') | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
2RRF
 
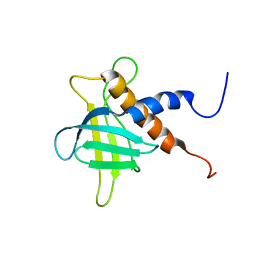 | | The solution structure of the C-terminal region of Zinc finger FYVE domain-containing protein 21 | | Descriptor: | Zinc finger FYVE domain-containing protein 21 | | Authors: | Koshiba, S, Tomizawa, T, Hayashi, F, Tochio, N, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-08-03 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ZF21 protein, a regulator of the disassembly of focal adhesions and cancer metastasis, contains a novel noncanonical pleckstrin homology domain
J.Biol.Chem., 286, 2011
|
|
3CPQ
 
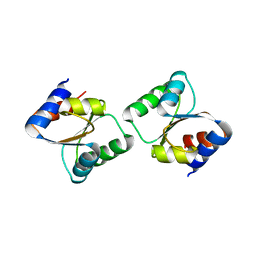 | | Crystal Structure of L30e a ribosomal protein from Methanocaldococcus jannaschii DSM2661 (MJ1044) | | Descriptor: | 50S ribosomal protein L30e | | Authors: | Jeyakanthan, J, Sarani, R, Mridula, P, Sekar, K, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of L30e a ribosomal protein from Methanocaldococcus jannaschii DSM2661 (MJ1044)
To be Published
|
|
3HPD
 
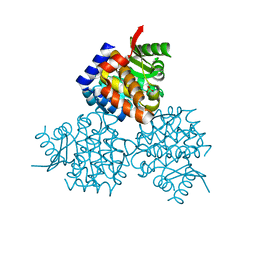 | | Structure of hydroxyethylthiazole kinase protein from pyrococcus horikoshii OT3 | | Descriptor: | Hydroxyethylthiazole kinase, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Thamotharan, S, Kuramitsu, S, Yokoyama, S, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-06-04 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Hydroxyethylthiazole Kinase Protein from Pyrococcus Horikoshii Ot3
To be Published
|
|
3GFJ
 
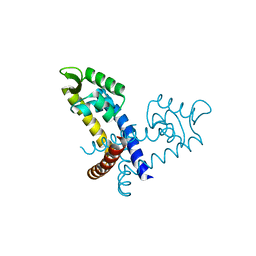 | |
3GFL
 
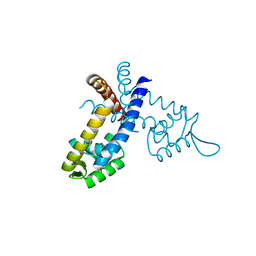 | |
3GEZ
 
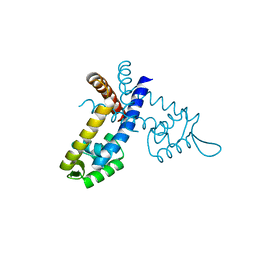 | |
