3KDH
 
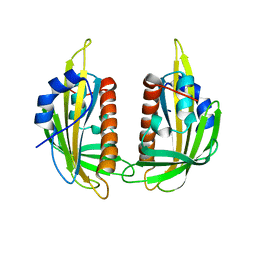 | | Structure of ligand-free PYL2 | | Descriptor: | Putative uncharacterized protein At2g26040 | | Authors: | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
3KDJ
 
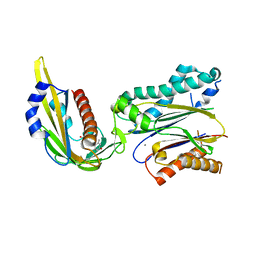 | | Complex structure of (+)-ABA-bound PYL1 and ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, MANGANESE (II) ION, Protein phosphatase 2C 56, ... | | Authors: | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | Deposit date: | 2009-10-23 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
3KDI
 
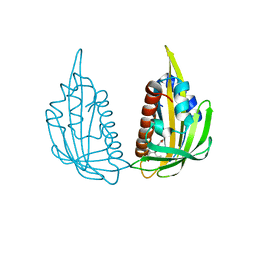 | | Structure of (+)-ABA bound PYL2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At2g26040 | | Authors: | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.379 Å) | | Cite: | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
4M59
 
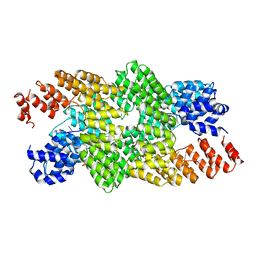 | | Crystal structure of the pentatricopeptide repeat protein PPR10 in complex with an 18-nt psaJ RNA element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
4GG4
 
 | | Crystal structure of the TAL effector dHax3 bound to specific DNA-RNA hybrid | | Descriptor: | DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3, RNA (5'-R(*AP*GP*AP*GP*AP*GP*AP*UP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3') | | Authors: | Yin, P, Deng, D, Yan, C.Y, Pan, X.J, Yan, N, Shi, Y.G. | | Deposit date: | 2012-08-05 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Specific DNA-RNA hybrid recognition by TAL effectors
Cell Rep, 2, 2012
|
|
4M57
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 from maize | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10 | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
5IWW
 
 | | Crystal structure of RNA editing factor of designer PLS-type PPR/9R protein in complex with MORF9/RIP9 | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic, PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
5IWB
 
 | | Crystal structure of design pentatricopeptide repeat complex with MORF protein | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic, PLS3-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-22 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structural basis for designer pentatricopeptide repeat proteins interaction with MORF protein
To be published
|
|
5WWD
 
 | | Crystal structure of AtNUDX1 | | Descriptor: | AMMONIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2016-12-31 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
5WY6
 
 | | Crystal structure of AtNUDX1 (E56A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nudix hydrolase 1, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2017-01-11 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
8Z9Z
 
 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
8Z9A
 
 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum bound with geranyl acetate | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
5IZW
 
 | | Crystal structure of RNA editing specific factor of designer PLS-type PPR-9R protein | | Descriptor: | PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-26 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
6L77
 
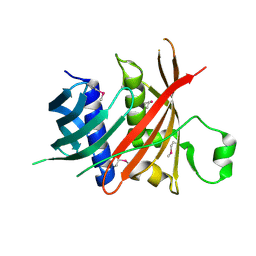 | | Crystal structure of MS5 from Brassica napus | | Descriptor: | MS5a | | Authors: | Wang, X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-10-31 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the meiosis-related protein MS5 reveals non-canonical papain enhancement by cystatin-like folds.
Febs Lett., 594, 2020
|
|
5JCB
 
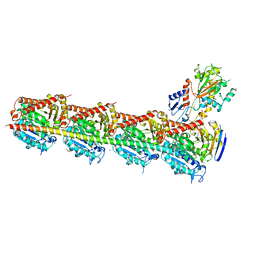 | | Microtubule depolymerizing agent podophyllotoxin derivative YJTSF1 | | Descriptor: | (5R,5aR,8aS,9R)-9-[(4H-1,2,4-triazol-3-yl)sulfanyl]-5-(3,4,5-trimethoxyphenyl)-5,8,8a,9-tetrahydro-2H-furo[3',4':6,7]naphtho[2,3-d][1,3]dioxol-6(5aH)-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Guan, Z, Zhao, W, Yin, P. | | Deposit date: | 2016-04-14 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Inhibition of Tubulin by the Antitumor Agent 4 beta-(1,2,4-triazol-3-ylthio)-4-deoxypodophyllotoxin.
ACS Chem. Biol., 12, 2017
|
|
7XC8
 
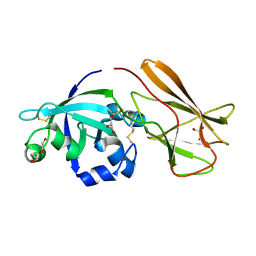 | | Crystal structure of cotton alpha-like expansin GhEXLA1 | | Descriptor: | Beta-expansin, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Zhao, F, Men, S, Xue, Y, Tu, L.L, Yin, P, Zhang, X.L. | | Deposit date: | 2022-03-23 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of cotton alpha-like expansin GhEXLA1
To Be Published
|
|
6LNI
 
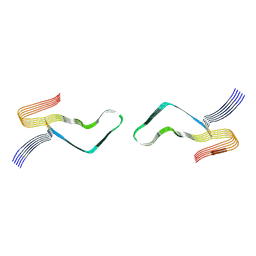 | | Cryo-EM structure of amyloid fibril formed by full-length human prion protein | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Wang, Q, Guan, Z.Y, Tao, J, Li, X.N, Hao, M.M, Chen, J, Zhang, D.L, Zhu, H.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2019-12-30 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.702 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human prion protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6M79
 
 | |
8GQE
 
 | | Crystal structure of the W285A mutant of UVR8 in complex with RUP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ultraviolet-B receptor UVR8, WD repeat-containing protein RUP2 | | Authors: | Wang, Y.D, Wang, L.X, Guan, Z.Y, chang, H.F, Yin, P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RUP2 facilitates UVR8 redimerization via two interfaces.
Plant Commun., 4, 2023
|
|
4DJK
 
 | | Structure of glutamate-GABA antiporter GadC | | Descriptor: | Probable glutamate/gamma-aminobutyrate antiporter | | Authors: | Ma, D, Lu, P.L, Yan, C.Y, Fan, C, Yin, P, Wang, J.W, Shi, Y.G. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structure and mechanism of a glutamate-GABA antiporter
Nature, 483, 2012
|
|
8W5J
 
 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8W5K
 
 | | Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
3U1M
 
 | | Structure of the mRNA splicing complex component Cwc2 | | Descriptor: | Pre-mRNA-splicing factor CWC2, ZINC ION | | Authors: | Lu, P, Lu, G, Yan, C, Wang, L, Li, W, Yin, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition
Biochem.J., 441, 2012
|
|
3U1L
 
 | | Structure of the mRNA splicing complex component Cwc2 | | Descriptor: | Pre-mRNA-splicing factor CWC2, ZINC ION | | Authors: | Lu, P, Lu, G, Yan, C, Wang, L, Li, W, Yin, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition
Biochem.J., 441, 2012
|
|
3NS2
 
 | | High-resolution structure of pyrabactin-bound PYL2 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Hao, Q, Yin, P, Yan, C, Yuan, X, Wang, J, Yan, N. | | Deposit date: | 2010-07-01 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | Single amino acid alteration between Valine and Isoleucine determines the distinct pyrabactin selectivity by PYL1 and PYL2
J.Biol.Chem., 285, 2010
|
|
