1S7M
 
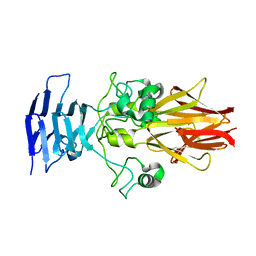 | | Crystal Structure of HiaBD1 | | Descriptor: | Hia | | Authors: | Yeo, H.J, Cotter, S.E, Laarmann, S, Juehne, T, St Geme, J.W, Waksman, G. | | Deposit date: | 2004-01-29 | | Release date: | 2004-04-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for host recognition by the Haemophilus influenzae Hia autotransporter.
Embo J., 23, 2004
|
|
1G6O
 
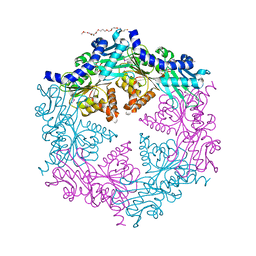 | | CRYSTAL STRUCTURE OF THE HELICOBACTER PYLORI ATPASE, HP0525, IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CAG-ALPHA, DI(HYDROXYETHYL)ETHER | | Authors: | Yeo, H.J, Savvides, S.N, Herr, A.B, Lanka, E, Waksman, G, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-11-07 | | Release date: | 2001-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system.
Mol.Cell, 6, 2000
|
|
1KA8
 
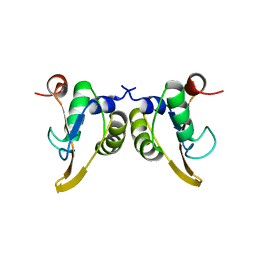 | | Crystal Structure of the Phage P4 Origin-Binding Domain | | Descriptor: | putative P4-specific DNA primase | | Authors: | Yeo, H.J, Ziegelin, G, Korolev, S, Calendar, R, Lanka, E, Waksman, G. | | Deposit date: | 2001-10-31 | | Release date: | 2002-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Phage P4 origin-binding domain structure reveals a mechanism for regulation of DNA-binding activity by homo- and heterodimerization of winged helix proteins.
Mol.Microbiol., 43, 2002
|
|
8OTX
 
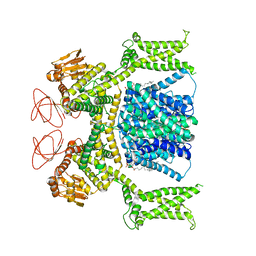 | | Cryo-EM structure of Strongylocentrotus purpuratus sperm-specific Na+/H+ exchanger SLC9C1 in nanodisc | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Sperm-specific sodium proton exchanger | | Authors: | Yeo, H, Mehta, V, Gulati, A, Drew, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure and electromechanical coupling of a voltage-gated Na + /H + exchanger.
Nature, 623, 2023
|
|
8OTW
 
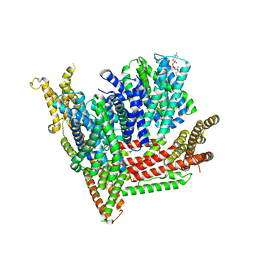 | | Cryo-EM structure of Strongylocentrotus purpuratus SLC9C1 in presence of cAMP | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sperm-specific sodium proton exchanger | | Authors: | Yeo, H, Mehta, V, Gulati, A, Drew, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structure and electromechanical coupling of a voltage-gated Na + /H + exchanger.
Nature, 623, 2023
|
|
8OTQ
 
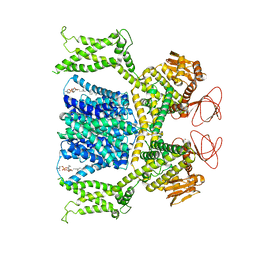 | | Cryo-EM structure of Strongylocentrotus purpuratus sperm-specific Na+/H+ exchanger SLC9C1 in GDN | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PALMITIC ACID, Sperm-specific sodium proton exchanger | | Authors: | Yeo, H, Mehta, V, Gulati, A, Drew, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structure and electromechanical coupling of a voltage-gated Na + /H + exchanger.
Nature, 623, 2023
|
|
3FS5
 
 | |
1R8I
 
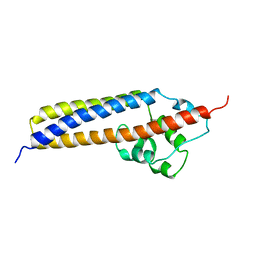 | | Crystal structure of TraC | | Descriptor: | TraC | | Authors: | Yeo, H.-J, Yuan, Q, Beck, M.R, Baron, C, Waksman, G. | | Deposit date: | 2003-10-24 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4O6J
 
 | | Crystal sturucture of T. acidophilum IdeR | | Descriptor: | FE (II) ION, Iron-dependent transcription repressor related protein | | Authors: | Lee, J.Y, Yeo, H.K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis and insight into metal-ion activation of the iron-dependent regulator from Thermoplasma acidophilum.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3D6X
 
 | | Crystal structure of Campylobacter jejuni FabZ | | Descriptor: | (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase | | Authors: | Yokoyama, T, Yeo, H.J. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Campylobacter jejuni fatty acid synthase II: structural and functional analysis of beta-hydroxyacyl-ACP dehydratase (FabZ).
Biochem.Biophys.Res.Commun., 380, 2009
|
|
3D6L
 
 | |
7RXU
 
 | | Crystal structure of Cj1090c | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Lipoprotein | | Authors: | Kim, Y, Yeo, H.J. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Campylobacter jejuni lipoprotein Cj1090c.
Proteins, 91, 2023
|
|
3BNV
 
 | |
5CJ9
 
 | | Bacillus halodurans Arginine repressor, ArgR | | Descriptor: | Arginine repressor, S-1,2-PROPANEDIOL | | Authors: | Park, Y.W, Kang, J, Yeo, H.Y, Lee, J.Y. | | Deposit date: | 2015-07-14 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Structural Analysis and Insights into the Oligomeric State of an Arginine-Dependent Transcriptional Regulator from Bacillus halodurans.
Plos One, 11, 2016
|
|
7CC7
 
 | |
3Q3I
 
 | |
4GIO
 
 | | Crystal structure of Campylobacter jejuni cj0090 | | Descriptor: | BROMIDE ION, Putative lipoprotein | | Authors: | Kawai, F, Yeo, H.J. | | Deposit date: | 2012-08-08 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Campylobacter jejuni Cj0090 protein reveals a novel variant of the immunoglobulin fold among bacterial lipoproteins.
Proteins, 80, 2012
|
|
3Q3H
 
 | |
5H3A
 
 | | Structural analysis of KSHV thymidylate synthase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ORF70, TOMUDEX | | Authors: | Choi, Y.M, Yeo, H.K, Park, Y.W, Lee, J.Y. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Thymidylate Synthase from Kaposi's Sarcoma-Associated Herpesvirus with the Anticancer Drug Raltitrexed.
PLoS ONE, 11, 2016
|
|
5GPC
 
 | | Structural analysis of fatty acid degradation regulator FadR from Bacillus halodurans | | Descriptor: | DNA (5'-D(P*CP*AP*TP*GP*AP*AP*TP*GP*AP*GP*TP*AP*TP*TP*CP*AP*TP*TP*CP*AP*T)-3'), DNA (5'-D(P*GP*AP*TP*GP*AP*AP*TP*GP*AP*AP*TP*AP*CP*TP*CP*AP*TP*TP*CP*AP*T)-3'), Transcriptional regulator (TetR/AcrR family) | | Authors: | Lee, J.Y, Yeo, H.K, Park, T.W. | | Deposit date: | 2016-08-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of operator sites recognition and effector binding in the TetR family transcription regulator FadR.
Nucleic Acids Res., 45, 2017
|
|
5GP9
 
 | | Structural analysis of fatty acid degradation regulator FadR from Bacillus halodurans | | Descriptor: | GLYCEROL, MAGNESIUM ION, PALMITIC ACID, ... | | Authors: | Lee, J.Y, Yeo, H.K, Park, Y.W. | | Deposit date: | 2016-08-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structural basis of operator sites recognition and effector binding in the TetR family transcription regulator FadR.
Nucleic Acids Res., 45, 2017
|
|
4O5V
 
 | | Crystal structure of T. acidophilum IdeR | | Descriptor: | FE (II) ION, Iron-dependent transcription repressor related protein | | Authors: | Lee, J.Y, Yeo, H.K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis and insight into metal-ion activation of the iron-dependent regulator from Thermoplasma acidophilum.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5H38
 
 | | Structural analysis of KSHV thymidylate synthase | | Descriptor: | ORF70, PHOSPHATE ION | | Authors: | Choi, Y.M, Yeo, H.K, Park, Y.W, Lee, J.Y. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of Thymidylate Synthase from Kaposi's Sarcoma-Associated Herpesvirus with the Anticancer Drug Raltitrexed.
PLoS ONE, 11, 2016
|
|
5GPA
 
 | |
5H39
 
 | | Structural analysis of KSHV thymidylate synthase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ORF70 | | Authors: | Choi, Y.M, Yeo, H.K, Park, Y.W, Lee, J.Y. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Thymidylate Synthase from Kaposi's Sarcoma-Associated Herpesvirus with the Anticancer Drug Raltitrexed.
PLoS ONE, 11, 2016
|
|
