5GAM
 
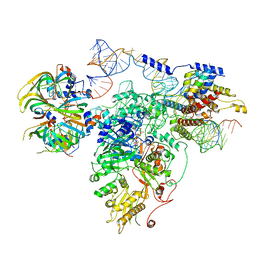 | | Foot region of the yeast spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Pre-mRNA-splicing factor 8, Pre-mRNA-splicing factor SNU114, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Angstrom resolution
Nature, 530, 2016
|
|
6KY1
 
 | |
6KY0
 
 | |
7BGB
 
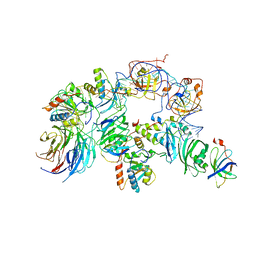 | | The H/ACA RNP lobe of human telomerase | | Descriptor: | H/ACA ribonucleoprotein complex subunit 1, H/ACA ribonucleoprotein complex subunit 2, H/ACA ribonucleoprotein complex subunit 3, ... | | Authors: | Nguyen, T.H.D, Ghanim, G.E, Fountain, A.J, van Roon, A.M.M, Rangan, R, Das, R, Collins, K. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human telomerase holoenzyme with bound telomeric DNA.
Nature, 593, 2021
|
|
7BG9
 
 | | The catalytic core lobe of human telomerase in complex with a telomeric DNA substrate | | Descriptor: | DNA (5'-D(P*TP*TP*AP*GP*GP*G)-3'), Histone H2A, Histone H2B, ... | | Authors: | Nguyen, T.H.D, Ghanim, G.E, Fountain, A.J, van Roon, A.M.M, Rangan, R, Das, R, Collins, K. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of human telomerase holoenzyme with bound telomeric DNA.
Nature, 593, 2021
|
|
5GAP
 
 | | Body region of the U4/U6.U5 tri-snRNP | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, Pre-mRNA-processing factor 31, Pre-mRNA-splicing factor 6, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Oubridge, C, Bai, X.C, Newman, A, Scheres, S, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
6LCI
 
 | | Solution structure of mdaA-1 domain | | Descriptor: | mdaA-1 | | Authors: | Nguyen, T.A, Le, S, Lee, M, Fan, J.S, Yang, D, Yan, J, Jedd, G. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Fungal Wound Healing through Instantaneous Protoplasmic Gelation.
Curr.Biol., 31, 2021
|
|
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6SX8
 
 | |
7KM4
 
 | |
4L56
 
 | | tRNA guanine transglycosylase H333D mutant apo structure | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Nguyen, T.X.P, Heine, A, Klebe, G. | | Deposit date: | 2013-06-10 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | What Glues a Homodimer Together: Systematic Analysis of the Stabilizing Effect of an Aromatic Hot Spot in the Protein-Protein Interface of the tRNA-Modifying Enzyme Tgt.
Acs Chem.Biol., 10, 2015
|
|
6T2A
 
 | |
6T6L
 
 | |
5FWZ
 
 | | Fasciola hepatica calcium binding protein FhCaBP2: Structure of the dynein light chain-like domain. P41212 mercury derivative. | | Descriptor: | CALCIUM BINDING PROTEIN, CHLORIDE ION, MERCURY (II) ION | | Authors: | Nguyen, T.H, Thomas, C.M, Timson, D.J, van Raaij, M.J. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fasciola hepatica calcium-binding protein FhCaBP2: structure of the dynein light chain-like domain.
Parasitol. Res., 115, 2016
|
|
1L5D
 
 | |
4IBN
 
 | | Crystal structure of LC9-RNase H1, a type 1 RNase H with the type 2 active-site motif | | Descriptor: | Ribonuclease H | | Authors: | Nguyen, T.-N, You, D.-J, Kanaya, E, Koga, Y, Kanaya, S. | | Deposit date: | 2012-12-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of metagenome-derived LC9-RNase H1 with atypical DEDN active site motif
Febs Lett., 587, 2013
|
|
4BGD
 
 | | Crystal structure of Brr2 in complex with the Jab1/MPN domain of Prp8 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nguyen, T.H.D, Li, J, Nagai, K. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Brr2-Prp8 Interactions and Implications for U5 Snrnp Biogenesis and the Spliceosome Active Site
Structure, 21, 2013
|
|
3U3G
 
 | | Structure of LC11-RNase H1 Isolated from Compost by Metagenomic Approach: Insight into the Structural Bases for Unusual Enzymatic Properties of Sto-RNase H1 | | Descriptor: | CHLORIDE ION, Ribonuclease H, UNKNOWN LIGAND | | Authors: | Nguyen, T.N, Angkawidjaja, C, Kanaya, E, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-10-05 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Activity, stability, and structure of metagenome-derived LC11-RNase H1, a homolog of Sulfolobus tokodaii RNase H1
Protein Sci., 21, 2012
|
|
1L5C
 
 | |
4H8K
 
 | | Crystal structure of LC11-RNase H1 in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3'), Ribonuclease H | | Authors: | Nguyen, T.N, You, D.J, Matsumoto, H, Kanaya, E, Kanaya, S. | | Deposit date: | 2012-09-23 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of metagenome-derived LC11-RNase H1 in complex with RNA/DNA hybrid
J.Struct.Biol., 182, 2013
|
|
