2QCU
 
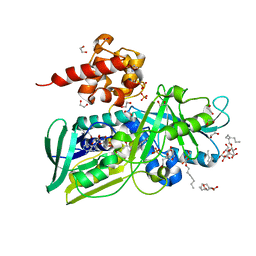 | | Crystal structure of Glycerol-3-phosphate Dehydrogenase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Aerobic glycerol-3-phosphate dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yeh, J.I, Chinte, U, Du, S. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
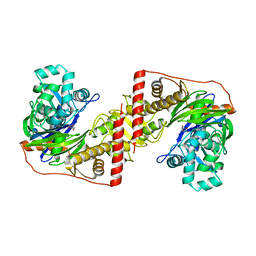
3D7E
 
 | |
1R59
 
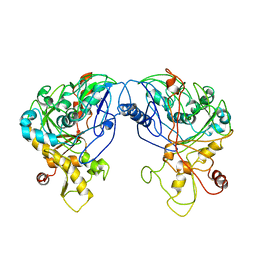 | | Enterococcus casseliflavus glycerol kinase | | Descriptor: | Glycerol kinase | | Authors: | Yeh, J.I, Charrier, V, Paulo, J, Hou, L, Darbon, E, Claiborne, A, Hol, W.G, Deutscher, J. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Enterococcal Glycerol Kinase in the Absence and Presence of Glycerol: Correlation of conformation to substrate binding and a mechanism of activation by phosphorylation
Biochemistry, 43, 2004
|
|
4E54
 
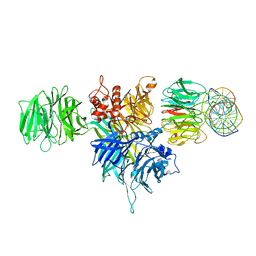 | |
4E5Z
 
 | |
3PA0
 
 | | Crystal Structure of Chiral Gamma-PNA with Complementary DNA Strand: Insight Into the Stability and Specificity of Recognition an Conformational Preorganization | | Descriptor: | 1,2-ETHANEDIOL, DNA 5'-D(*AP*TP*CP*TP*GP*TP*GP*GP*TP*C)-3', PEPTIDE NUCLEIC ACID, ... | | Authors: | Yeh, J.I, Shivachev, B, Du, S. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of chiral gammaPNA with complementary DNA strand: insights into the stability and specificity of recognition and conformational preorganization.
J.Am.Chem.Soc., 132, 2010
|
|
1F8W
 
 | |
1M5W
 
 | | 1.96 A Crystal Structure of Pyridoxine 5'-Phosphate Synthase in Complex with 1-deoxy-D-xylulose phosphate | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, PHOSPHATE ION, Pyridoxal phosphate biosynthetic protein pdxJ | | Authors: | Yeh, J.I, Du, S, Pohl, E, Cane, D.E. | | Deposit date: | 2002-07-10 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multistate Binding in Pyridoxine 5'-Phosphate Synthase: 1.96 A Crystal Structure in
Complex with 1-deoxy-D-xylulose phosphate
Biochemistry, 41, 2002
|
|
2R4J
 
 | | Crystal structure of Escherichia coli SeMet substituted Glycerol-3-phosphate Dehydrogenase in complex with DHAP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, Aerobic glycerol-3-phosphate dehydrogenase, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-31 | | Release date: | 2008-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2R46
 
 | | Crystal structure of Escherichia coli Glycerol-3-phosphate Dehydrogenase in complex with 2-phosphopyruvic acid. | | Descriptor: | 1,2-ETHANEDIOL, Aerobic glycerol-3-phosphate dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-30 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential
monotopic membrane enzyme involved in respiration and metabolism
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2R45
 
 | | Crystal structure of Escherichia coli Glycerol-3-phosphate Dehydrogenase in complex with 2-phospho-d-glyceric acid | | Descriptor: | 1,2-ETHANEDIOL, 2-PHOSPHOGLYCERIC ACID, Aerobic glycerol-3-phosphate dehydrogenase, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-30 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential
monotopic membrane enzyme involved in respiration and metabolism
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2R4E
 
 | | Crystal structure of Escherichia coli Glycerol-3-phosphate Dehydrogenase in complex with DHAP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, Aerobic glycerol-3-phosphate dehydrogenase, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-31 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1JOA
 
 | | NADH PEROXIDASE WITH CYSTEINE-SULFENIC ACID | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH PEROXIDASE | | Authors: | Yeh, J.I, Claiborne, A.C, Hol, W.G.J. | | Deposit date: | 1996-07-02 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the native cysteine-sulfenic acid redox center of enterococcal NADH peroxidase refined at 2.8 A resolution.
Biochemistry, 35, 1996
|
|
1XUP
 
 | | ENTEROCOCCUS CASSELIFLAVUS GLYCEROL KINASE COMPLEXED WITH GLYCEROL | | Descriptor: | GLYCEROL, Glycerol kinase | | Authors: | Yeh, J.I, Charrier, V, Paulo, J, Hou, L, Darbon, E, Hol, W.G.J, Deutscher, J. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of Enterococcal Glycerol Kinase in the Absence and Presence of Glycerol: Correlation of Conformation to Substrate Binding and a Mechanism of Activation by Phosphorylation
Biochemistry, 43, 2004
|
|
3H3N
 
 | | Glycerol Kinase H232R with Glycerol | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glycerol kinase, ... | | Authors: | Yeh, J.I, Kettering, R.D. | | Deposit date: | 2009-04-16 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural characterizations of glycerol kinase: unraveling phosphorylation-induced long-range activation
Biochemistry, 48, 2009
|
|
3H46
 
 | | Glycerol Kinase H232E with Glycerol | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glycerol kinase, ... | | Authors: | Yeh, J.I, Kettering, R.D. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterizations of glycerol kinase: unraveling phosphorylation-induced long-range activation
Biochemistry, 48, 2009
|
|
3H3O
 
 | | Glycerol Kinase H232R with Ethylene Glycol | | Descriptor: | 1,2-ETHANEDIOL, Glycerol kinase, PHOSPHATE ION | | Authors: | Yeh, J.I, Kettering, R.D. | | Deposit date: | 2009-04-16 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterizations of glycerol kinase: unraveling phosphorylation-induced long-range activation
Biochemistry, 48, 2009
|
|
3MBS
 
 | | Crystal structure of 8mer PNA | | Descriptor: | 1,2-ETHANEDIOL, Peptide Nucleic Acid | | Authors: | Yeh, J.I, Pohl, E, Truan, D, He, W, Sheldrick, G.M, Achim, C. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | The crystal structure of non-modified and bipyridine-modified PNA duplexes.
Chemistry, 16, 2010
|
|
3H45
 
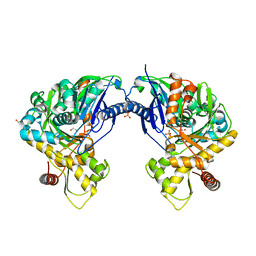 | | Glycerol Kinase H232E with Ethylene Glycol | | Descriptor: | 1,2-ETHANEDIOL, Glycerol kinase, PHOSPHATE ION | | Authors: | Yeh, J.I, Kettering, R.D. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural characterizations of glycerol kinase: unraveling phosphorylation-induced long-range activation
Biochemistry, 48, 2009
|
|
3MBU
 
 | | Structure of a bipyridine-modified PNA duplex | | Descriptor: | 1,2-ETHANEDIOL, Bipyridine-PNA, CARBONATE ION, ... | | Authors: | Yeh, J.I, Pohl, E, Truan, D, He, W, Sheldrick, G.M, Du, S, Achim, C. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of non-modified and bipyridine-modified PNA duplexes.
Chemistry, 16, 2010
|
|
2LIG
 
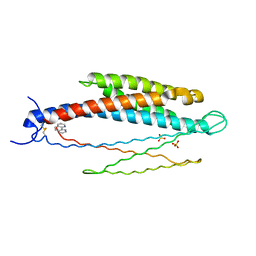 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR, ASPARTIC ACID, ... | | Authors: | Kim, S.-H, Yeh, J.I, Prive, G.G, Milburn, M, Scott, W, Koshland Junior, D.E. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
4U5W
 
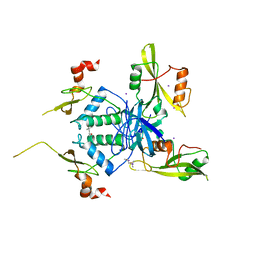 | | Crystal Structure of HIV-1 Nef-SF2 Core Domain in Complex with the Src Family Kinase Hck SH3-SH2 Tandem Regulatory Domains | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IODIDE ION, Protein Nef, ... | | Authors: | Alvarado, J.J, Yeh, J.I, Smithgall, T.E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Interaction with the Src Homology (SH3-SH2) Region of the Src-family Kinase Hck Structures the HIV-1 Nef Dimer for Kinase Activation and Effector Recruitment.
J.Biol.Chem., 289, 2014
|
|
1LIH
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Scott, W, Yeh, J.I, Prive, G.G, Milburn, M. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
1VLS
 
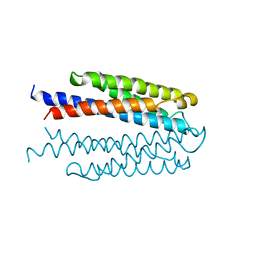 | | LIGAND BINDING DOMAIN OF THE WILD-TYPE ASPARTATE RECEPTOR | | Descriptor: | ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Yeh, J.I, Biemann, H.-P, Prive, G, Pandit, J, Koshland Junior, D.E. | | Deposit date: | 1996-09-17 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor.
J.Mol.Biol., 262, 1996
|
|
1VLT
 
 | | LIGAND BINDING DOMAIN OF THE WILD-TYPE ASPARTATE RECEPTOR WITH ASPARTATE | | Descriptor: | ASPARTATE RECEPTOR, ASPARTIC ACID | | Authors: | Kim, S.-H, Yeh, J.I, Biemann, H.-P, Prive, G, Pandit, J, Koshland Junior, D.E. | | Deposit date: | 1996-09-17 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor.
J.Mol.Biol., 262, 1996
|
|
