6COQ
 
 | | CSP1-K6A | | 分子名称: | Competence-stimulating peptide type 1 | | 著者 | Yang, Y. | | 登録日 | 2018-03-12 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COP
 
 | | CSP1-R3A | | 分子名称: | Competence-stimulating peptide type 1 | | 著者 | Yang, Y. | | 登録日 | 2018-03-12 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
5ZHQ
 
 | |
6COT
 
 | | CSP2-d10 | | 分子名称: | Competence-stimulating peptide type 2 | | 著者 | Yang, Y. | | 登録日 | 2018-03-12 | | 公開日 | 2018-08-29 | | 最終更新日 | 2018-09-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6DUG
 
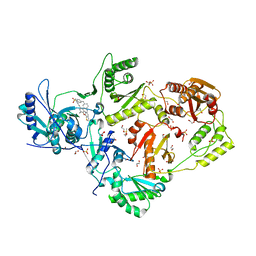 | | Crystal structure of HIV-1 reverse transcriptase K101P mutant in complex with non-nucleoside inhibitor 25a | | 分子名称: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | 著者 | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | 登録日 | 2018-06-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.225 Å) | | 主引用文献 | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6J0L
 
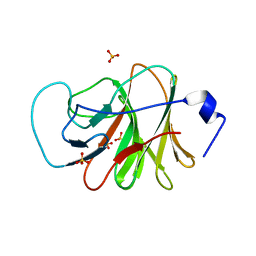 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with sulfate ion | | 分子名称: | Butyrophilin subfamily 3 member A3, SULFATE ION | | 著者 | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | 登録日 | 2018-12-24 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
2JOB
 
 | | Solution structure of an antilipopolysaccharide factor from shrimp and its possible Lipid A binding site | | 分子名称: | antilipopolysaccharide factor | | 著者 | Yang, Y, Boze, H, Chemardin, P, Padilla, A, Moulin, G, Tassanakajon, A, Pugniere, M, Roquet, F, Gueguen, Y, Bachere, E, Aumelas, A. | | 登録日 | 2007-03-02 | | 公開日 | 2008-03-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of rALF-Pm3, an anti-lipopolysaccharide factor from shrimp: Model of the possible lipid A-binding site
Biopolymers, 91, 2009
|
|
2MIU
 
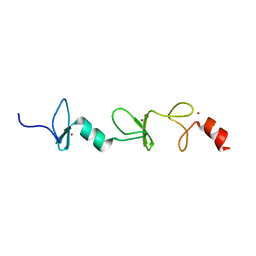 | | Structure of FHL2 LIM adaptor and its Interaction with Ski | | 分子名称: | Four and a half LIM domains protein 2, ZINC ION | | 著者 | Yang, Y, Sun, Y, Medrano, E.E, Tian, X, Weiss, M.A. | | 登録日 | 2013-12-20 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of FHL2 LIM adaptor and its Interaction with Ski
To be Published
|
|
2M9K
 
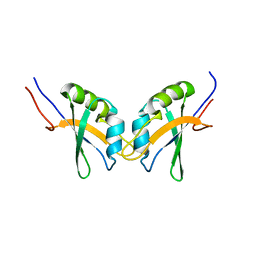 | | RBPMS2-Nter | | 分子名称: | RNA-binding protein with multiple splicing 2 | | 著者 | Yang, Y. | | 登録日 | 2013-06-12 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Homodimerization of RBPMS2 through a new RRM-interaction motif is necessary to control smooth muscle plasticity.
Nucleic Acids Res., 42, 2014
|
|
6CD3
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142A from Cupriavidus metallidurans in complex with 3-HAA | | 分子名称: | 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | 著者 | Yang, Y, Liu, F, Liu, A. | | 登録日 | 2018-02-07 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.612 Å) | | 主引用文献 | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6BVQ
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase N27A from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | 著者 | Yang, Y, Liu, F, Liu, A. | | 登録日 | 2017-12-13 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.084 Å) | | 主引用文献 | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6BVP
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase N27A from Cupriavidus metallidurans | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | 著者 | Yang, Y, Liu, F, Liu, A. | | 登録日 | 2017-12-13 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.903 Å) | | 主引用文献 | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6BVR
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142A from Cupriavidus metallidurans | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | 著者 | Yang, Y, Liu, F, Liu, A. | | 登録日 | 2017-12-13 | | 公開日 | 2018-06-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6D62
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142P from Cupriavidus metallidurans in complex with 3-HAA | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | 著者 | Yang, Y, Liu, F, Liu, A. | | 登録日 | 2018-04-19 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6D60
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142P from Cupriavidus metallidurans | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | 著者 | Yang, Y, Liu, F, Liu, A. | | 登録日 | 2018-04-19 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6BVS
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142A from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | 著者 | Yang, Y, Liu, F, Liu, A. | | 登録日 | 2017-12-13 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.318 Å) | | 主引用文献 | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6D61
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142P from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | 著者 | Yang, Y, Liu, F, Liu, A. | | 登録日 | 2018-04-19 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
4NLH
 
 | |
6CSD
 
 | | V308E mutant of cytochrome P450 2D6 complexed with prinomastat | | 分子名称: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Yang, Y.T, Fujita, K, Wang, P.F, Im, S.C, Pearl, N.M, Meagher, J, Stuckey, J, Waskell, L. | | 登録日 | 2018-03-20 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.391 Å) | | 主引用文献 | Characteristic conformational changes on the distal and proximal surfaces of cytochrome P450 2D6 in response to substrate binding
To Be Published
|
|
4MGJ
 
 | | Crystal structure of cytochrome P450 2B4 F429H in complex with 4-CPI | | 分子名称: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Yang, Y, Zhang, H, Usharani, D, Bu, W, Im, S, Tarasev, M, Rwere, F, Meagher, J, Sun, C, Stuckey, J, Shaik, S, Waskell, L. | | 登録日 | 2013-08-28 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural and Functional Characterization of a Cytochrome P450 2B4 F429H Mutant with an Axial Thiolate-Histidine Hydrogen Bond.
Biochemistry, 53, 2014
|
|
5ZZ3
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A3 | | 分子名称: | Butyrophilin, subfamily 3, member A3 isoform b variant | | 著者 | Yang, Y.Y, Li, X, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | 登録日 | 2018-05-30 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
5ZXK
 
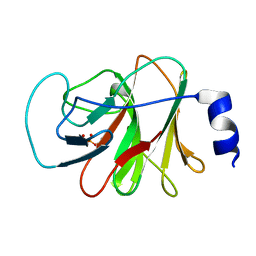 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with HMBPP | | 分子名称: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A1 | | 著者 | Yang, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | 登録日 | 2018-05-21 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
2JZ1
 
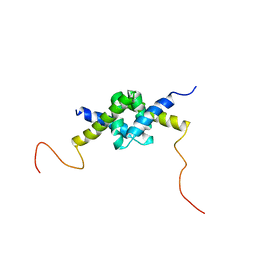 | | DSX_long | | 分子名称: | Protein doublesex | | 著者 | Yang, Y, Zhang, W, Bayrer, J.R, Weiss, M.A. | | 登録日 | 2007-12-21 | | 公開日 | 2008-01-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Doublesex and the Regulation of Sexual Dimorphism in Drosophila melanogaster: STRUCTURE, FUNCTION, AND MUTAGENESIS OF A FEMALE-SPECIFIC DOMAIN.
J.Biol.Chem., 283, 2008
|
|
2KMM
 
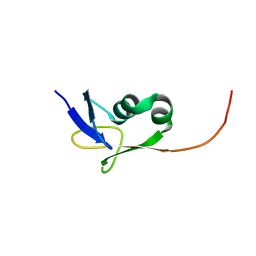 | | Solution NMR structure of the TGS domain of PG1808 from Porphyromonas gingivalis. Northeast Structural Genomics Consortium Target PgR122A (418-481) | | 分子名称: | Guanosine-3',5'-bis(Diphosphate) 3'-pyrophosphohydrolase | | 著者 | Yang, Y, Ramelot, T.A, Cort, J.R, Lee, D, Ciccosanti, C, Hamilton, K, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-07-31 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the TGS domain of PG1808 from
Porphyromonas gingivalis. Northeast Structural Genomics Consortium Target
PgR122A (418-481)
To be Published
|
|
2K6H
 
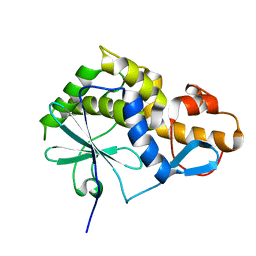 | |
