4WD8
 
 | |
4WD7
 
 | |
5OCV
 
 | | A Rare Lysozyme Crystal Form Solved Using High-Redundancy 3D Electron Diffraction Data from Micron-Sized Needle Shaped Crystals | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Xu, H, Lebrette, H, Yang, T, Srinivas, V, Hovmoller, S, Hogbom, M, Zou, X. | | Deposit date: | 2017-07-03 | | Release date: | 2018-03-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.2 Å) | | Cite: | A Rare Lysozyme Crystal Form Solved Using Highly Redundant Multiple Electron Diffraction Datasets from Micron-Sized Crystals.
Structure, 26, 2018
|
|
7YG7
 
 | | Structure of the Spring Viraemia of Carp Virus ribonucleoprotein Complex | | Descriptor: | Nucleoprotein, RNA (99-mer) | | Authors: | Liu, B, Wang, Z.X, Yang, T, Yu, D.Q, Ouyang, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Spring Viraemia of Carp Virus Ribonucleoprotein Complex Reveals Its Assembly Mechanism and Application in Antiviral Drug Screening.
J.Virol., 97, 2023
|
|
7X1Z
 
 | | Structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
7X1Y
 
 | | Structure of the phosphorylation-site double mutant S431A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
7DCY
 
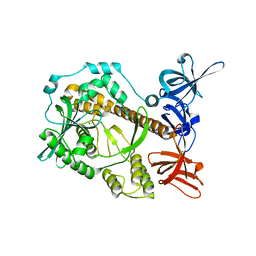 | | Apo form of Mycoplasma genitalium RNase R | | Descriptor: | MAGNESIUM ION, Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DIC
 
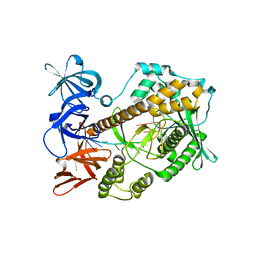 | | Mycoplasma genitalium RNase R in complex with single-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DOL
 
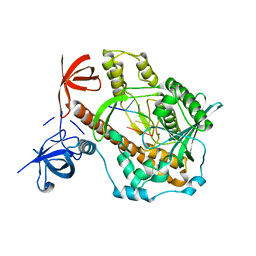 | | Mycoplasma genitalium RNase R in complex with double-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DID
 
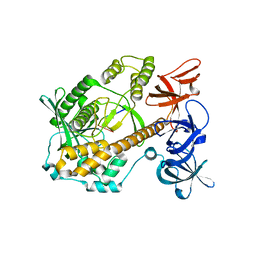 | | Mycoplasma genitalium RNase R in complex with ribose methylated single-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
8ECY
 
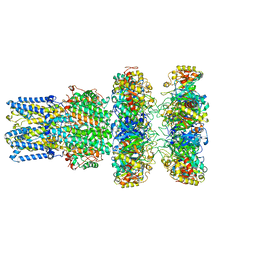 | | cryoEM structure of bovine bestrophin-2 and glutamine synthetase complex | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Owji, A.P, Kittredge, A.K, Yang, T. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Bestrophin-2 and glutamine synthetase form a complex for glutamate release.
Nature, 611, 2022
|
|
6IHR
 
 | |
6IHS
 
 | |
6IHL
 
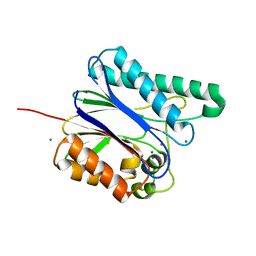 | | Crystal structure of bacterial serine phosphatase | | Descriptor: | MAGNESIUM ION, Phosphorylated protein phosphatase | | Authors: | Yang, C.-G, yang, T. | | Deposit date: | 2018-09-30 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Structural Insight into the Mechanism of Staphylococcus aureus Stp1 Phosphatase.
Acs Infect Dis., 5, 2019
|
|
6IHW
 
 | |
6IHU
 
 | |
6IHV
 
 | |
6IHT
 
 | |
7DFT
 
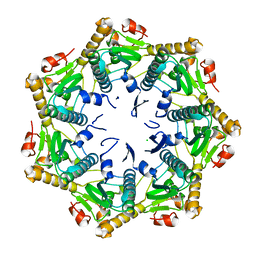 | | Crystal structure of Xanthomonas oryzae ClpP | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CHLORIDE ION | | Authors: | Yang, C.-G, Yang, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dysregulation of ClpP by Small-Molecule Activators Used Against Xanthomonas oryzae pv. oryzae Infections.
J.Agric.Food Chem., 69, 2021
|
|
7DFU
 
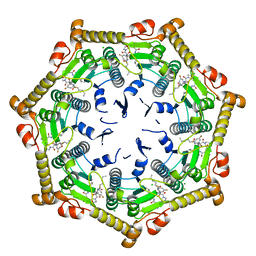 | |
1ZVM
 
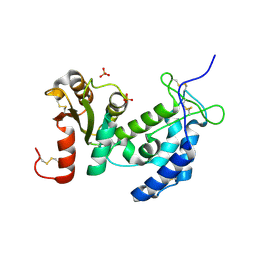 | | Crystal structure of human CD38: cyclic-ADP-ribosyl synthetase/NAD+ glycohydrolase | | Descriptor: | ADP-ribosyl cyclase 1, SULFATE ION | | Authors: | Shi, W, Yang, T, Almo, S.C, Schramm, V.L, Sauve, A. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human CD38: Cyclic-ADP-ribosyl synthetase/NAD+ glycohydrolase
To be Published
|
|
9DYK
 
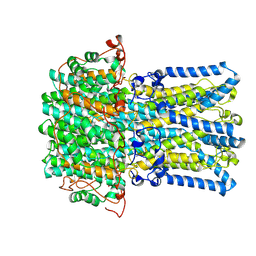 | | BEST2 + GABA closed state | | Descriptor: | Bestrophin-2, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-10-14 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Neurotransmitter-bound bestrophin channel structures reveal small molecule drug targeting sites for disease treatment.
Nat Commun, 15, 2024
|
|
9DYL
 
 | | BEST1 + PABA open state | | Descriptor: | 4-AMINOBENZOIC ACID, Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-10-14 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Neurotransmitter-bound bestrophin channel structures reveal small molecule drug targeting sites for disease treatment.
Nat Commun, 15, 2024
|
|
9DYM
 
 | | BEST1 + PABA intermediate state | | Descriptor: | 4-AMINOBENZOIC ACID, Bestrophin-1, CALCIUM ION, ... | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-10-14 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Neurotransmitter-bound bestrophin channel structures reveal small molecule drug targeting sites for disease treatment.
Nat Commun, 15, 2024
|
|
9DYN
 
 | | BEST2 + PABA open state | | Descriptor: | 4-AMINOBENZOIC ACID, Bestrophin-2a, CALCIUM ION, ... | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-10-14 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Neurotransmitter-bound bestrophin channel structures reveal small molecule drug targeting sites for disease treatment.
Nat Commun, 15, 2024
|
|
