9BRT
 
 | | Intact V-ATPase State 1 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRA
 
 | | Intact V-ATPase State 2 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRR
 
 | | Intact V-ATPase State 3 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRS
 
 | | Intact V-ATPase State 2 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRY
 
 | | V0-only V-ATPase in synaptophysin gene knock-out mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRQ
 
 | | Intact V-ATPase State 3 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
2HBX
 
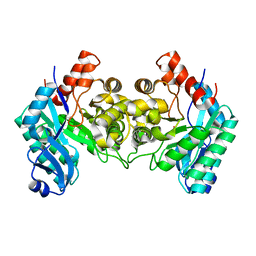 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
2HBV
 
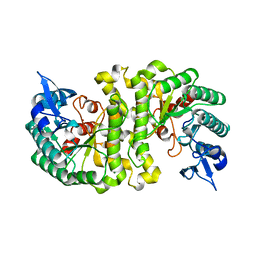 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
4ICY
 
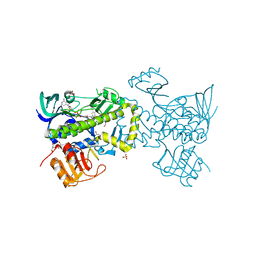 | | Tracing the Evolution of Angucyclinone Monooxygenases: Structural Determinants for C-12b Hydroxylation and Substrate Inhibition in PgaE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kallio, P, Patrikainen, P, Belogurov, G, Mantsala, P, Yang, K, Niemi, J, Metsa-Ketela, M. | | Deposit date: | 2012-12-11 | | Release date: | 2013-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tracing the Evolution of Angucyclinone Monooxygenases: Structural Determinants for C-12b Hydroxylation and Substrate Inhibition in PgaE.
Biochemistry, 52, 2013
|
|
4KJJ
 
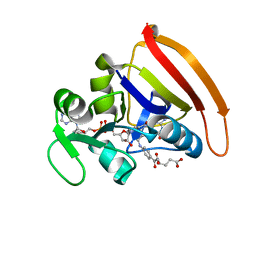 | | Cryogenic WT DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
4KJL
 
 | | Room Temperature N23PPS148A DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
4KJK
 
 | | Room Temperature WT DHFR | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
1RKU
 
 | | Crystal Structure of ThrH gene product of Pseudomonas Aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, homoserine kinase | | Authors: | Singh, S.K, Yang, K, Subramanian, K, Karthikeyan, S, Huynh, T, Zhang, X, Phillips, M.A, Zhang, H. | | Deposit date: | 2003-11-23 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The thrH Gene Product of Pseudomonas aeruginosa Is a Dual Activity Enzyme with a Novel Phosphoserine:Homoserine Phosphotransferase Activity.
J.Biol.Chem., 279, 2004
|
|
1S17
 
 | | Identification of Novel Potent Bicyclic Peptide Deformylase Inhibitors | | Descriptor: | 2-(3,4-DIHYDRO-3-OXO-2H-BENZO[B][1,4]THIAZIN-2-YL)-N-HYDROXYACETAMIDE, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Molteni, V, He, X, Nabakka, J, Yang, K, Kreusch, A, Gordon, P, Bursulaya, B, Ryder, N.S, Goldberg, R, He, Y. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of novel potent bicyclic peptide deformylase inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1RKV
 
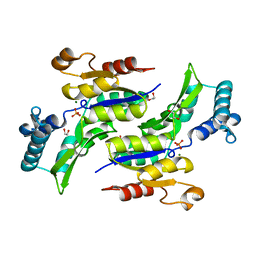 | | Structure of Phosphate complex of ThrH from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Singh, S.K, Yang, K, Subramanian, K, Karthikeyan, S, Huynh, T, Zhang, X, Phillips, M.A, Zhang, H. | | Deposit date: | 2003-11-23 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The thrH Gene Product of Pseudomonas aeruginosa Is a Dual Activity Enzyme with a Novel Phosphoserine:Homoserine Phosphotransferase Activity.
J.Biol.Chem., 279, 2004
|
|
1WNO
 
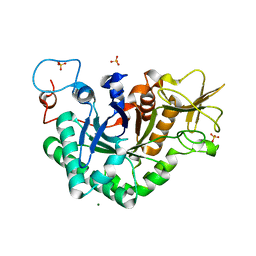 | | Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Hu, H, Wang, G, Yang, H, Zhou, J, Mo, L, Yang, K, Jin, C, Jin, C, Rao, Z. | | Deposit date: | 2004-08-07 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407
To be Published
|
|
1WOF
 
 | | Crystal Structure Of SARS-CoV Mpro in Complex with an Inhibitor N1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]-L-ALANYL-L-VALYL-N~1~-((1S)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Yang, H, Bartlam, M, Xue, X, Yang, K, Liang, W, Rao, Z. | | Deposit date: | 2004-08-18 | | Release date: | 2005-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
