7XUF
 
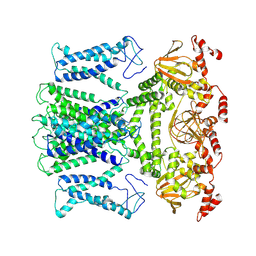 | | Cryo-EM structure of the AKT1-AtKC1 complex from Arabidopsis thaliana | | Descriptor: | POTASSIUM ION, Potassium channel AKT1, Potassium channel KAT3 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Yang, F, Zhang, Y.M, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7WSW
 
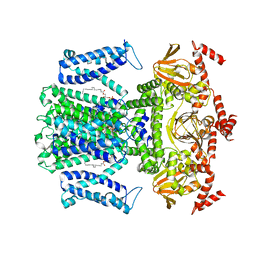 | | Cryo-EM structure of the Potassium channel AKT1 from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
8JD9
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
7VR1
 
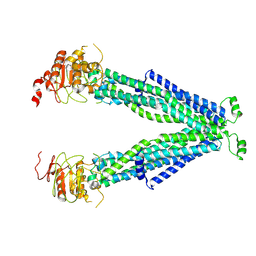 | |
8JDA
 
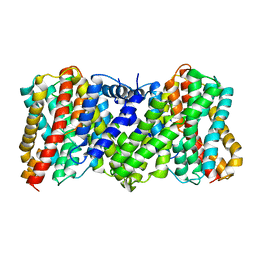 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2 | | Descriptor: | Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
7FCV
 
 | | Cryo-EM structure of the Potassium channel AKT1 mutant from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Zhang, Y.M, Tang, R.F, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7CQ7
 
 | | Structure of the human CLCN7-OSTM1 complex with ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex with ADP
To Be Published
|
|
7CQ5
 
 | | Structure of the human CLCN7-OSTM1 complex with ATP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex with ATP
To Be Published
|
|
7CQ6
 
 | | Structure of the human CLCN7-OSTM1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, H(+)/Cl(-) exchange transporter 7, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex
To Be Published
|
|
5FN4
 
 | | Cryo-EM structure of gamma secretase in class 2 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FN5
 
 | | Cryo-EM structure of gamma secretase in class 3 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FN2
 
 | | Cryo-EM structure of gamma secretase in complex with a drug DAPT | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FN3
 
 | | Cryo-EM structure of gamma secretase in class 1 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5XLS
 
 | | Crystal structure of UraA in occluded conformation | | Descriptor: | 12-TUNGSTOPHOSPHATE, URACIL, Uracil permease | | Authors: | Yu, X.Z, Yang, G.H, Yan, C.Y, Yan, N. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric structure of the uracil:proton symporter UraA provides mechanistic insights into the SLC4/23/26 transporters
Cell Res., 27, 2017
|
|
7D3B
 
 | | flavone reductase | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, Cd1, FLAVIN MONONUCLEOTIDE | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7D3A
 
 | | flavone reductase | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Cd1, FLAVIN MONONUCLEOTIDE | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7D39
 
 | | FLR-apo | | Descriptor: | Cd1, FLAVIN MONONUCLEOTIDE | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7D38
 
 | | flavone reductase | | Descriptor: | Cd1, FLAVIN MONONUCLEOTIDE, chrysin | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
