6KNB
 
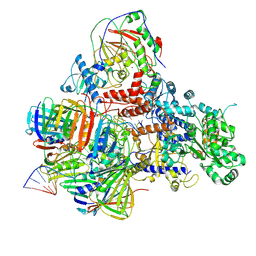 | | PolD-PCNA-DNA (form A) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
1DQ3
 
 | |
1RDB
 
 | |
1RDC
 
 | |
1RDD
 
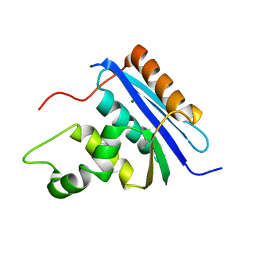 | |
1RDA
 
 | |
2RN2
 
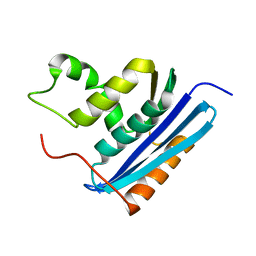 | | STRUCTURAL DETAILS OF RIBONUCLEASE H FROM ESCHERICHIA COLI AS REFINED TO AN ATOMIC RESOLUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Katayanagi, K, Miyagawa, M, Matsushima, M, Ishikawa, M, Kanaya, S, Nakamura, H, Ikehara, M, Matsuzaki, T, Morikawa, K. | | Deposit date: | 1992-04-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural details of ribonuclease H from Escherichia coli as refined to an atomic resolution.
J.Mol.Biol., 223, 1992
|
|
6KNC
 
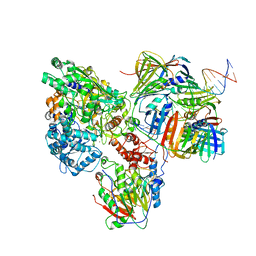 | | PolD-PCNA-DNA (form B) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
2D0K
 
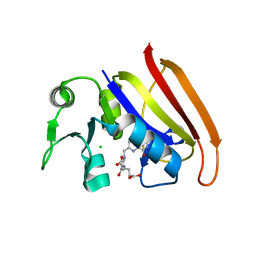 | |
1FUT
 
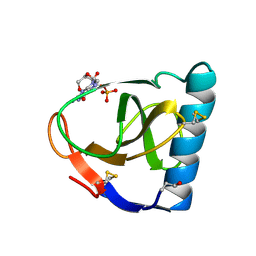 | | CRYSTAL STRUCTURES OF RIBONUCLEASE F1 OF FUSARIUM MONILIFORME IN ITS FREE FORM AND IN COMPLEX WITH 2'GMP | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE F1 | | Authors: | Katayanagi, K, Vassylyev, D.G, Ishikawa, K, Morikawa, K. | | Deposit date: | 1993-01-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of ribonuclease F1 of Fusarium moniliforme in its free form and in complex with 2'GMP.
J.Mol.Biol., 230, 1993
|
|
1FUS
 
 | |
1D49
 
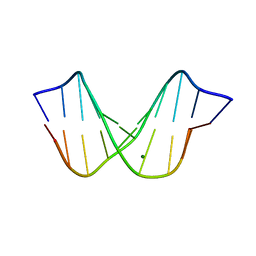 | | THE STRUCTURE OF A B-DNA DECAMER WITH A CENTRAL T-A STEP: C-G-A-T-T-A-A-T-C-G | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*AP*AP*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Grzeskowiak, K, Yanagi, K, Dickerson, R.E. | | Deposit date: | 1991-09-17 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G.
J.Mol.Biol., 225, 1992
|
|
1DA3
 
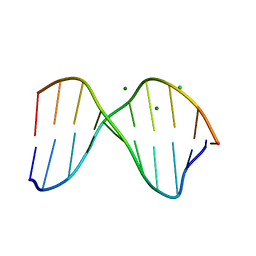 | | THE CRYSTAL STRUCTURE OF THE TRIGONAL DECAMER C-G-A-T-C-G-6MEA-T-C-G: A B-DNA HELIX WITH 10.6 BASE-PAIRS PER TURN | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*AP*TP*CP*GP*(6MA)P*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Baikalov, I, Grzeskowiak, K, Yanagi, K, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-11-09 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the trigonal decamer C-G-A-T-C-G-6meA-T-C-G: a B-DNA helix with 10.6 base-pairs per turn.
J.Mol.Biol., 231, 1993
|
|
5DNB
 
 | |
1D23
 
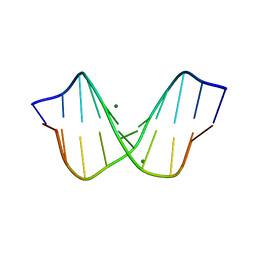 | | THE STRUCTURE OF B-HELICAL C-G-A-T-C-G-A-T-C-G AND COMPARISON WITH C-C-A-A-C-G-T-T-G-G. THE EFFECT OF BASE PAIR REVERSALS | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*AP*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Grzeskowiak, K, Yanagi, K, Prive, G.G, Dickerson, R.E. | | Deposit date: | 1991-05-29 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of B-helical C-G-A-T-C-G-A-T-C-G and comparison with C-C-A-A-C-G-T-T-G-G. The effect of base pair reversals.
J.Biol.Chem., 266, 1991
|
|
1V29
 
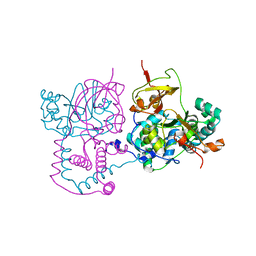 | | Crystal structure of Nitrile hydratase from a thermophile Bacillus smithii | | Descriptor: | COBALT (II) ION, nitrile hydratase a chain, nitrile hydratase b chain | | Authors: | Hourai, S, Miki, M, Takashima, Y, Mitsuda, S, Yanagi, K. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of nitrile hydratase from a thermophilic Bacillus smithii
Biochem.Biophys.Res.Commun., 312, 2003
|
|
3ANT
 
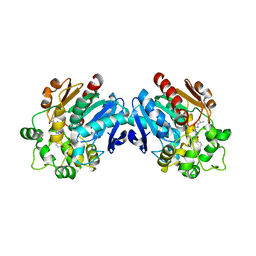 | | Human soluble epoxide hydrolase in complex with a synthetic inhibitor | | Descriptor: | 4-[3-(1-methylethyl)-1,2,4-oxadiazol-5-yl]-N-[(1S,2R)-2-phenylcyclopropyl]piperidine-1-carboxamide, Epoxide hydrolase 2 | | Authors: | Chiyo, N, Ishii, T, Hourai, S, Yanagi, K. | | Deposit date: | 2010-09-08 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A practical use of ligand efficiency indices out of the fragment-based approach: ligand efficiency-guided lead identification of soluble epoxide hydrolase inhibitors
J.Med.Chem., 54, 2011
|
|
3ANS
 
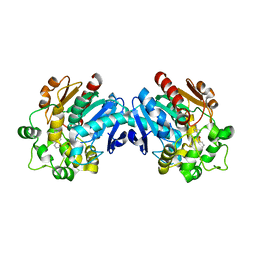 | | Human soluble epoxide hydrolase in complex with a synthetic inhibitor | | Descriptor: | 4-cyano-N-[(1S,2R)-2-phenylcyclopropyl]benzamide, Epoxide hydrolase 2 | | Authors: | Chiyo, N, Ishii, T, Hourai, S, Yanagi, K. | | Deposit date: | 2010-09-08 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A practical use of ligand efficiency indices out of the fragment-based approach: ligand efficiency-guided lead identification of soluble epoxide hydrolase inhibitors
J.Med.Chem., 54, 2011
|
|
1KVB
 
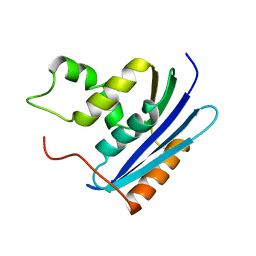 | | E. COLI RIBONUCLEASE HI D134H MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVA
 
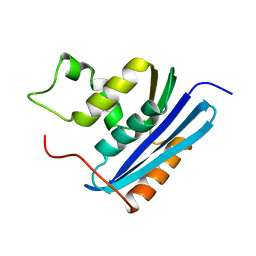 | | E. COLI RIBONUCLEASE HI D134A MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1ENK
 
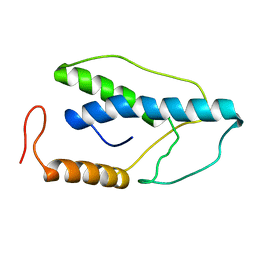 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1ENI
 
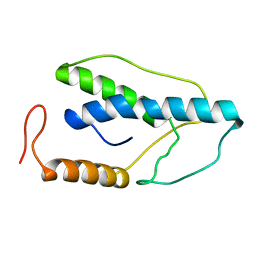 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1ENJ
 
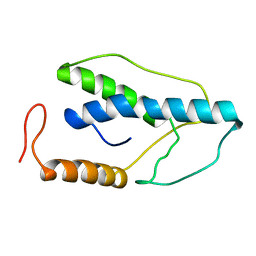 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1KVC
 
 | | E. COLI RIBONUCLEASE HI D134N MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
3A57
 
 | | Crystal structure of Thermostable Direct Hemolysin | | Descriptor: | Thermostable direct hemolysin 2 | | Authors: | Hashimoto, H, Yanagihara, I, Nakahira, K, Hamada, D, Ikegami, T, Mayanagi, K, Kaieda, S, Fukui, T, Ohnishi, K, Kajiyama, S, Yamane, T, Ikeguchi, M, Honda, T, Shimizu, T, Sato, M. | | Deposit date: | 2009-08-03 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and functional characterization of Vibrio parahaemolyticus thermostable direct hemolysin
J.Biol.Chem., 285, 2010
|
|
