4WD7
 
 | |
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
4WD8
 
 | |
8YFE
 
 | | Cryo EM structure of Komagataella phaffii Rat1-Rai1 complex | | Descriptor: | 5'-3' exoribonuclease, Decapping nuclease | | Authors: | Yanagisawa, T, Murayama, Y, Ehara, H, Sekine, S.I. | | Deposit date: | 2024-02-24 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
8YF5
 
 | | Cryo EM structure of Komagataella phaffii Rat1-Rai1-Rtt103 complex | | Descriptor: | 5'-3' exoribonuclease, Decapping nuclease, Exonuclease Rat1p and Rai1p interacting protein | | Authors: | Yanagisawa, T, Murayama, Y, Ehara, H, Sekine, S.I. | | Deposit date: | 2024-02-24 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
3VQV
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with AMPPNP (re-refined) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pyrrolysine--tRNA ligase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VQW
 
 | | Crystal structure of the SeMet substituted catalytic domain of pyrrolysyl-tRNA synthetase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pyrrolysine--tRNA ligase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VQY
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with BocLys and AMPPNP (form 2) | | Descriptor: | MAGNESIUM ION, N~6~-(tert-butoxycarbonyl)-L-lysine, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-02 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VQX
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in triclinic crystal form | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, Pyrrolysine--tRNA ligase, ... | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-02 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WNC
 
 | | Crystal structure of EF-Pyl in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
3WNB
 
 | | Crystal structure of EF-Pyl in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
3WND
 
 | | Crystal structure of EF-Pyl | | Descriptor: | CITRIC ACID, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
8IFJ
 
 | |
6JP2
 
 | |
2E3C
 
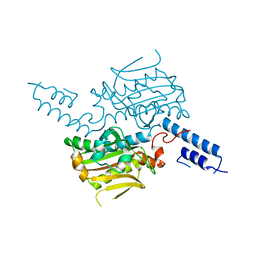 | |
2EL7
 
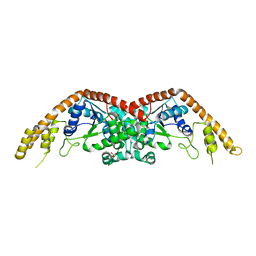 | | Crystal structure of Tryptophanyl-tRNA synthetase from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, Tryptophanyl-tRNA synthetase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Thermus thermophilus
To be published
|
|
2ZIN
 
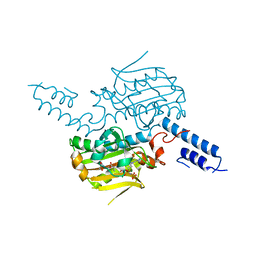 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with BocLys and an ATP analogue | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N~6~-(tert-butoxycarbonyl)-L-lysine, ... | | Authors: | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-19 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode N(varepsilon)-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification
Chem.Biol., 15, 2008
|
|
2ZCE
 
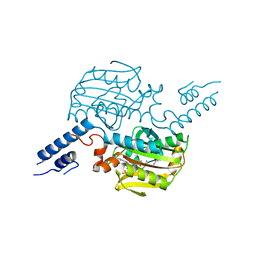 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with pyrrolysine and an ATP analogue | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PYRROLYSINE, ... | | Authors: | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Studies on Multiple Conformational States of Active-site Loops in Pyrrolysyl-tRNA Synthetase
J.Mol.Biol., 378, 2008
|
|
2ZIO
 
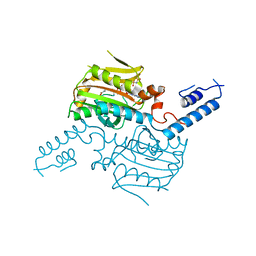 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with AlocLys-AMP and PNP | | Descriptor: | 5'-O-[(S)-({(2S)-2-amino-6-[(propoxycarbonyl)amino]hexanoyl}oxy)(hydroxy)phosphoryl]adenosine, IMIDODIPHOSPHORIC ACID, Pyrrolysyl-tRNA synthetase | | Authors: | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-19 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode N(varepsilon)-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification
Chem.Biol., 15, 2008
|
|
2YVL
 
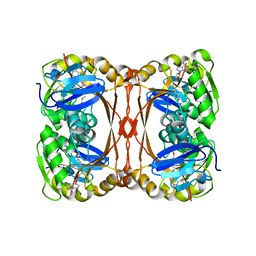 | |
6ABK
 
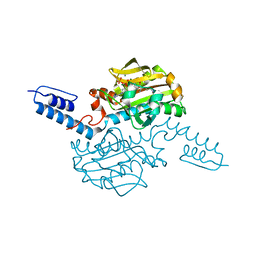 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with TeocLys | | Descriptor: | (2S)-2-azanyl-6-(trimethylsilylmethoxycarbonylamino)hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-22 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAP
 
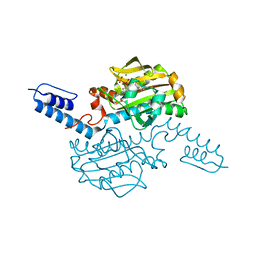 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with ZaeSeCys | | Descriptor: | 3-[(2-{[(benzyloxy)carbonyl]amino}ethyl)selanyl]-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAO
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with TCO*Lys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.403 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AB0
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with pAmPyLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N6-{[(6-aminopyridin-3-yl)methoxy]carbonyl}-L-lysine, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAC
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with mAzZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
