6LZJ
 
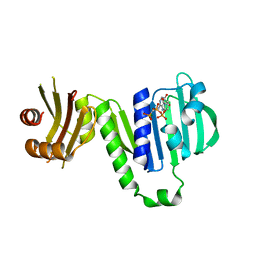 | | Aquifex aeolicus MutL ATPase domain complexed with AMPPCP | | Descriptor: | DNA mismatch repair protein MutL, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Fukui, K, Izuhara, K, Yano, T. | | Deposit date: | 2020-02-19 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72835565 Å) | | Cite: | A Lynch syndrome-associated mutation at a Bergerat ATP-binding fold destabilizes the structure of the DNA mismatch repair endonuclease MutL.
J.Biol.Chem., 295, 2020
|
|
4RZE
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, L437W,D594E mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Fengwei, L, Xuyang, T, Anzhong, L, Li, L, Yuan, L, Hangzi, C, Qiao, W, Tianwei, L. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
6VX7
 
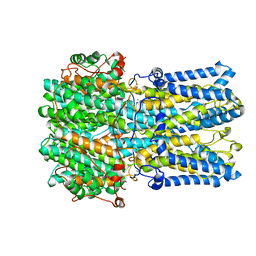 | | bestrophin-2 Ca2+-bound state (5 mM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX9
 
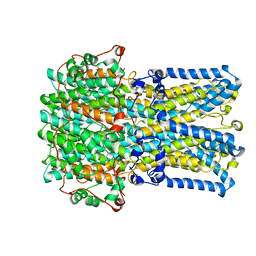 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX8
 
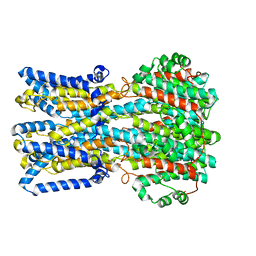 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX6
 
 | | bestrophin-2 Ca2+-bound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX5
 
 | | bestrophin-2 Ca2+- unbound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5OCV
 
 | | A Rare Lysozyme Crystal Form Solved Using High-Redundancy 3D Electron Diffraction Data from Micron-Sized Needle Shaped Crystals | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Xu, H, Lebrette, H, Yang, T, Srinivas, V, Hovmoller, S, Hogbom, M, Zou, X. | | Deposit date: | 2017-07-03 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.2 Å) | | Cite: | A Rare Lysozyme Crystal Form Solved Using Highly Redundant Multiple Electron Diffraction Datasets from Micron-Sized Crystals.
Structure, 26, 2018
|
|
2VPX
 
 | | Polysulfide reductase with bound quinone (UQ1) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VPZ
 
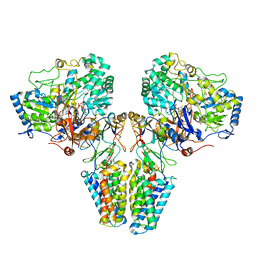 | | Polysulfide reductase native structure | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VPY
 
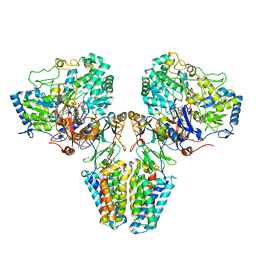 | | Polysulfide reductase with bound quinone inhibitor, pentachlorophenol (PCP) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VPW
 
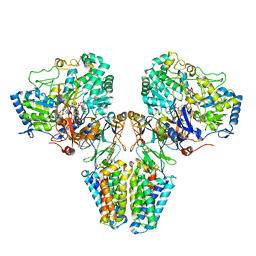 | | Polysulfide reductase with bound menaquinone | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8YFQ
 
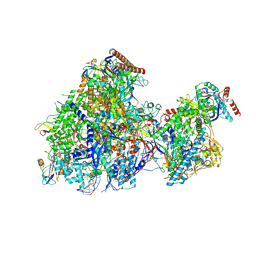 | | Cryo EM structure of Komagataella phaffii RNAPII-Rat1-Rai1 pre-termination complex | | Descriptor: | 5'-3' exoribonuclease, DNA (90-mer), DNA-directed RNA polymerase subunit, ... | | Authors: | Murayama, Y, Yanagisawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2024-02-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
8YFR
 
 | | Cryo EM structure of Komagataella phaffii Rat1-Rai1 complex bound within the RNAPII cleft | | Descriptor: | 5'-3' exoribonuclease, DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Murayama, Y, Yanagisawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2024-02-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
9CTS
 
 | | Best1 + GABA intermediate state 2 | | Descriptor: | Bestrophin-1, CALCIUM ION, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-07-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | GAD65 tunes the functions of Best1 as a GABA receptor and a neurotransmitter conducting channel.
Nat Commun, 15, 2024
|
|
9CTQ
 
 | | Best1 + GABA open state | | Descriptor: | Bestrophin-1, CALCIUM ION, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-07-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | GAD65 tunes the functions of Best1 as a GABA receptor and a neurotransmitter conducting channel.
Nat Commun, 15, 2024
|
|
9CTT
 
 | | Best1 + GABA closed state | | Descriptor: | Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-07-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | GAD65 tunes the functions of Best1 as a GABA receptor and a neurotransmitter conducting channel.
Nat Commun, 15, 2024
|
|
9CTR
 
 | | Best1 + GABA intermediate state 1 | | Descriptor: | Bestrophin-1, CALCIUM ION, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-07-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | GAD65 tunes the functions of Best1 as a GABA receptor and a neurotransmitter conducting channel.
Nat Commun, 15, 2024
|
|
8YQ9
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS V1/S) complexed with FB6, NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-[4-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]oxypropoxy]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D, Saeyang, T, Arwon, U, Hoarau, M, Decharuangsilp, S, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel flexible biphenyl Pf DHFR inhibitors with improved antimalarial activity.
Rsc Med Chem, 15, 2024
|
|
8YQ8
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS V1/S) complexed with FB8, NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-[4-[4-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]oxybutoxy]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D, Saeyang, T, Arwon, U, Hoarau, M, Decharuangsilp, S, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel flexible biphenyl Pf DHFR inhibitors with improved antimalarial activity.
Rsc Med Chem, 15, 2024
|
|
3N36
 
 | | Erythrina corallodendron lectin mutant (Y106G) in complex with Galactose | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Thamotharan, S, Karthikeyan, T, Kulkarni, K.A, Shetty, K.N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2010-05-19 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modification of the sugar specificity of a plant lectin: structural studies on a point mutant of Erythrina corallodendron lectin.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3N3H
 
 | | Erythrina corallodendron lectin mutant (Y106G) in complex with citrate | | Descriptor: | CALCIUM ION, CITRIC ACID, Lectin, ... | | Authors: | Thamotharan, S, Karthikeyan, T, Kulkarni, K.A, Shetty, K.N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2010-05-20 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modification of the sugar specificity of a plant lectin: structural studies on a point mutant of Erythrina corallodendron lectin.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3N35
 
 | | Erythrina corallodendron lectin mutant (Y106G) with N-Acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, Lectin, ... | | Authors: | Thamotharan, S, Karthikeyan, T, Kulkarni, K.A, Shetty, K.N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2010-05-19 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modification of the sugar specificity of a plant lectin: structural studies on a point mutant of Erythrina corallodendron lectin.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6LZI
 
 | | Aquifex aeolicus MutL ATPase domain complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL, ... | | Authors: | Fukui, K, Izuhara, K, Yano, T. | | Deposit date: | 2020-02-19 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69105685 Å) | | Cite: | A Lynch syndrome-associated mutation at a Bergerat ATP-binding fold destabilizes the structure of the DNA mismatch repair endonuclease MutL.
J.Biol.Chem., 295, 2020
|
|
6LZK
 
 | | Aquifex aeolicus MutL ATPase domain with K252N mutation | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL, SODIUM ION | | Authors: | Fukui, K, Izuhara, K, Yano, T. | | Deposit date: | 2020-02-19 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15949631 Å) | | Cite: | A Lynch syndrome-associated mutation at a Bergerat ATP-binding fold destabilizes the structure of the DNA mismatch repair endonuclease MutL.
J.Biol.Chem., 295, 2020
|
|
