5XC2
 
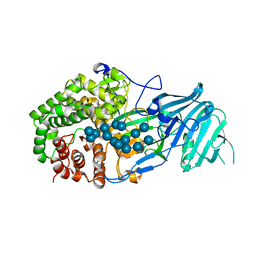 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminarihexaose | | Descriptor: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Qin, Z, Yang, S, Peng, Z, Yan, Q, Jiang, Z. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
5XWF
 
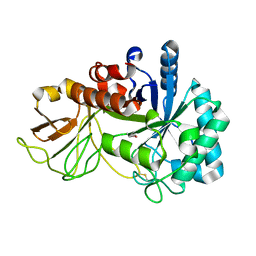 | | Crystal structure of chitinase (RmChi1) from Rhizomucor miehei (SP3221/SAD) | | Descriptor: | Fungal chitinase from Rhizomucor miehei (SeMet-substituted proteins) | | Authors: | Jiang, Z.Q, Hu, S.Q, Liu, Y.C, Qin, Z, Yan, Q.J, Yang, S.Q. | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.581 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 2021
|
|
5JVV
 
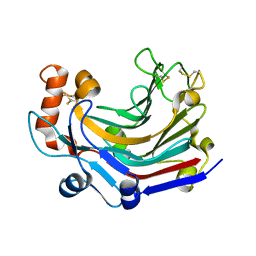 | | Crystal structure and characterization an elongating GH family 16 beta-1,3-glucosyltransferase | | Descriptor: | beta-1,3-glucosyltransferase | | Authors: | Qin, Z, Yan, Q, Yang, S, Jiang, Z. | | Deposit date: | 2016-05-11 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Catalytic Mechanism of a Novel Glycoside Hydrolase Family 16 "Elongating" beta-Transglycosylase
J. Biol. Chem., 292, 2017
|
|
4NRS
 
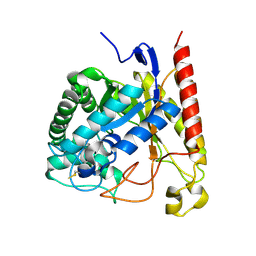 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NRR
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-beta-D-fructofuranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3ECN
 
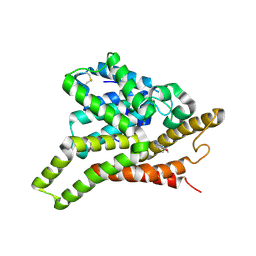 | | Crystal structure of PDE8A catalytic domain in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
3ECM
 
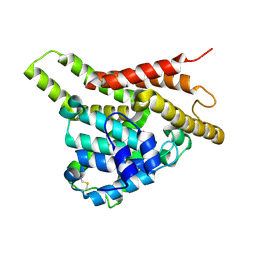 | | Crystal structure of the unliganded PDE8A catalytic domain | | Descriptor: | High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ZINC ION | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
4K35
 
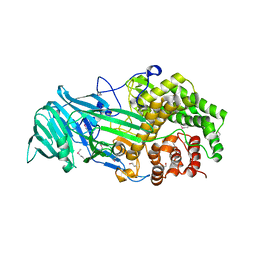 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | Authors: | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4LYP
 
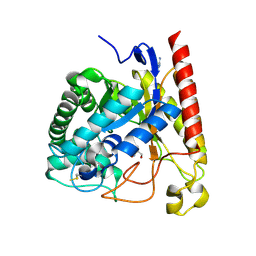 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Exo-beta-1,4-mannosidase, GUANIDINE | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5H9Y
 
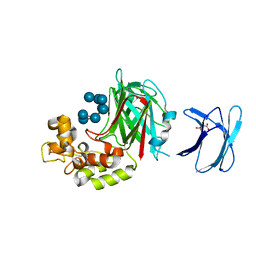 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii complexed with laminarihexaose. | | Descriptor: | L(+)-TARTARIC ACID, beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | Deposit date: | 2015-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
5H9X
 
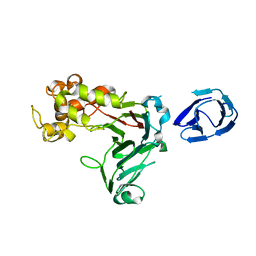 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii | | Descriptor: | beta-1,3-glucanase | | Authors: | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | Deposit date: | 2015-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
4LYQ
 
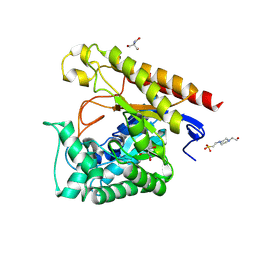 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E202A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase, ... | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LYR
 
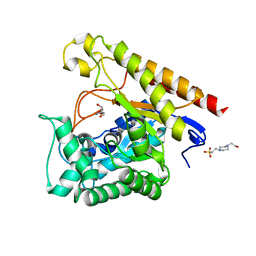 | | Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E301A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5YUQ
 
 | | The high resolution structure of chitinase (RmChi1) from the thermophilic fungus Rhizomucor miehei (sp P1) | | Descriptor: | Chintase | | Authors: | Jiang, Z.Q, Hu, S.Q, Liu, Y.C, Qin, Z, Yan, Q.J, Yang, S.Q. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 2021
|
|
4K3A
 
 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | Descriptor: | SULFATE ION, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | Authors: | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2FKJ
 
 | | The crystal structure of engineered OspA | | Descriptor: | Outer Surface Protein A | | Authors: | Makabe, K, Terechko, V, Gawlak, G, Yan, S, Koide, S. | | Deposit date: | 2006-01-04 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Atomic structures of peptide self-assembly mimics.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4EAI
 
 | | Co-crystal structure of an AMPK core with AMP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EAK
 
 | | Co-crystal structure of an AMPK core with ATP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EAJ
 
 | | Co-crystal of AMPK core with AMP soaked with ATP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
7RS9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-25 | | Descriptor: | (1S,2R,4S)-N-[4-(benzyloxy)phenyl]-5,6-bis(4-hydroxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RRY
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-20 | | Descriptor: | (1S,2R,4S,5S,6S)-5,6-bis(4-hydroxyphenyl)-N-{4-[3-(piperidin-1-yl)propoxy]phenyl}-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonamide, CHLORIDE ION, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RRZ
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-30 | | Descriptor: | (1S,2R,4S,5R,6S)-5-(4-hydroxyphenyl)-N-(4-methoxyphenyl)-6-{4-[3-(piperidin-1-yl)propoxy]phenyl}-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS0
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-18 | | Descriptor: | (1R,2S,4R,5R,6R)-5-(4-hydroxyphenyl)-N-(4-methoxyphenyl)-6-(4-propoxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS4
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-8 | | Descriptor: | (2E)-3-{4-[(1E)-2-(2-chloro-4-fluorophenyl)-1-(2H-indazol-5-yl)but-1-en-1-yl]phenyl}prop-2-enoic acid, CHLORIDE ION, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS2
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-23 | | Descriptor: | (2E)-3-(4-{[(1S,2R,4S,5S,6S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonyl](2,2,2-trifluoroethyl)amino}phenyl)prop-2-enoic acid, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
