1HSW
 
 | |
1EUI
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Authors: | Ravishankar, R, Sagar, M.B, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray analysis of a complex of Escherichia coli uracil DNA glycosylase (EcUDG) with a proteinaceous inhibitor. The structure elucidation of a prokaryotic UDG.
Nucleic Acids Res., 26, 1998
|
|
1HSX
 
 | |
2PEL
 
 | | PEANUT LECTIN | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PEANUT LECTIN, ... | | Authors: | Banerjee, R, Das, K, Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1995-08-23 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Conformation, protein-carbohydrate interactions and a novel subunit association in the refined structure of peanut lectin-lactose complex.
J.Mol.Biol., 259, 1996
|
|
5BOH
 
 | |
7VVI
 
 | | OXA-58 crystal structure of acylated meropenem complex | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, SULFATE ION | | Authors: | Saino, H, Sugiyabu, T, Miyano, M. | | Deposit date: | 2021-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | OXA-58 crystal structure of acylated meropenem complex
to be published
|
|
1XEK
 
 | |
1XEI
 
 | |
1XEJ
 
 | |
7OBZ
 
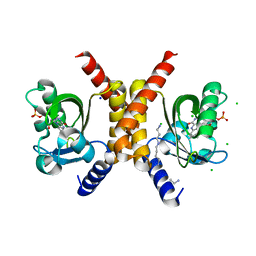 | | Structure of RsLOV D109G | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, LOV protein, ... | | Authors: | Moeglich, A, Krafft, T.G.A, Weyand, M. | | Deposit date: | 2021-04-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Light-Oxygen-Voltage Receptor Integrates Light and Temperature.
J.Mol.Biol., 433, 2021
|
|
7OB0
 
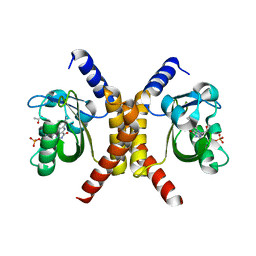 | | Structure of RsLOV d2 variant | | Descriptor: | ACETATE ION, CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Moeglich, A, Krafft, T.G.A, Weyand, M. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Light-Oxygen-Voltage Receptor Integrates Light and Temperature.
J.Mol.Biol., 433, 2021
|
|
1J3B
 
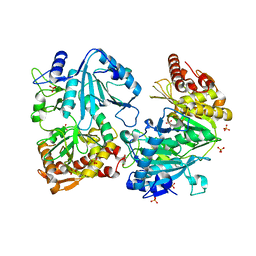 | | Crystal structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 | | Descriptor: | ATP-dependent phosphoenolpyruvate carboxykinase, CALCIUM ION, GLYCEROL, ... | | Authors: | Sugahara, M, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-21 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 showing the structural basis of induced fit and thermostability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1F10
 
 | |
3AFP
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | Descriptor: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A5U
 
 | | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex | | Descriptor: | DNA (31-MER), Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Manjunath, G.P, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2009-08-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex.
To be Published, 2009
|
|
1QUV
 
 | |
1GYM
 
 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C IN COMPLEX WITH GLUCOSAMINE-(ALPHA-1-6)-MYO-INOSITOL | | Descriptor: | (1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W, Ryan, M, Smith, M.P, Weaver, L.H, Keana, J.F.W, Griffith, O.H. | | Deposit date: | 1996-05-02 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with glucosaminyl(alpha 1-->6)-D-myo-inositol, an essential fragment of GPI anchors.
Biochemistry, 35, 1996
|
|
5XLW
 
 | | Mycobacterium tuberculosis Pantothenate kinase mutant F247A/F254A | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Paul, A, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Biochemical and structural studies of mutants indicate concerted movement of the dimer interface and ligand-binding region of Mycobacterium tuberculosis pantothenate kinase
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1ODK
 
 | |
1OD6
 
 | | The Crystal Structure of Phosphopantetheine adenylyltransferase from Thermus Thermophilus in complex with 4'-phosphopantetheine | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Takahashi, H, Inagaki, E, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Implications for the Thermal Stability of Phosphopantetheine Adenylyltransferase from Thermus Thermophilus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5XMB
 
 | | Mycobacterium tuberculosis Pantothenate kinase mutant F247A | | Descriptor: | Pantothenate kinase, SULFATE ION | | Authors: | Paul, A, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2017-05-13 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical and structural studies of mutants indicate concerted movement of the dimer interface and ligand-binding region of Mycobacterium tuberculosis pantothenate kinase
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5XLV
 
 | | Mycobacterium tuberculosis Pantothenate kinase mutant F254A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pantothenate kinase, ... | | Authors: | Paul, A, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural studies of mutants indicate concerted movement of the dimer interface and ligand-binding region of Mycobacterium tuberculosis pantothenate kinase
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1F0W
 
 | | CRYSTAL STRUCTURE OF ORTHORHOMBIC LYSOZYME GROWN AT PH 6.5 | | Descriptor: | LYSOZYME | | Authors: | Biswal, B.K, Sukumar, N, Vijayan, M. | | Deposit date: | 2000-05-17 | | Release date: | 2000-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hydration, mobility and accessibility of lysozyme: structures of a pH 6.5 orthorhombic form and its low-humidity variant and a comparative study involving 20 crystallographically independent molecules.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1F88
 
 | | CRYSTAL STRUCTURE OF BOVINE RHODOPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, RETINAL, ... | | Authors: | Okada, T, Palczewski, K, Stenkamp, R.E, Miyano, M. | | Deposit date: | 2000-06-29 | | Release date: | 2000-08-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of rhodopsin: A G protein-coupled receptor.
Science, 289, 2000
|
|
7VYN
 
 | | Matrix arm of active state CI from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-14 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
