4BN5
 
 | | Structure of human SIRT3 in complex with SRT1720 inhibitor | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLYCEROL, N-{2-[3-(piperazin-1-ylmethyl)imidazo[2,1-b][1,3]thiazol-6-yl]phenyl}quinoxaline-2-carboxamide, ... | | Authors: | Nguyen, G.T.T, Schaefer, S, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-05-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of Human Sirtuin 3 Complexes with Adp-Ribose and with Carba-Nad+ and Srt1720: Binding Details and Inhibition Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BVB
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH THE INHIBITOR EX-527 AND ADP-RIBOSE | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-3, MITOCHONDRIAL, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1WWC
 
 | | NT3 BINDING DOMAIN OF HUMAN TRKC RECEPTOR | | Descriptor: | PROTEIN (NT-3 GROWTH FACTOR RECEPTOR TRKC) | | Authors: | Ultsch, M.H, Wiesmann, C, Simmons, L.C, Henrich, J, Yang, M, Reilly, D, Bass, S.H, De Vos, A.M. | | Deposit date: | 1999-04-30 | | Release date: | 1999-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC.
J.Mol.Biol., 290, 1999
|
|
4BVE
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH THIOALKYLIMIDATE FORMED FROM THIO-ACETYL-LYSINE ACS2-PEPTIDE | | Descriptor: | 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, MITOCHONDRIAL, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BVH
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH THE INHIBITOR EX-527 AND 2'-O-ACETYL-ADP-RIBOSE | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, 1,2-ETHANEDIOL, 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BV3
 
 | | CRYSTAL STRUCTURE OF SIRT3 IN COMPLEX WITH THE INHIBITOR EX-527 AND NAD | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, ... | | Authors: | Gertz, M, Nguyen, N.T.T, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BVF
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH THIOALKYLIMIDATE FORMED FROM THIO-ACETYL-LYSINE ACS2-PEPTIDE CRYSTALLIZED IN PRESENCE OF THE INHIBITOR EX-527 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, MITOCHONDRIAL, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5YEI
 
 | | Mechanistic insight into the regulation of Pseudomonas aeruginosa aspartate kinase | | Descriptor: | Aspartokinase, GLYCEROL, LYSINE, ... | | Authors: | Li, C, Yang, M, Liu, L, Peng, C, Li, T, He, L, Song, Y, Zhu, Y, Bao, R. | | Deposit date: | 2017-09-17 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the allosteric regulation of Pseudomonas aeruginosa aspartate kinase.
Biochem.J., 475, 2018
|
|
4CLF
 
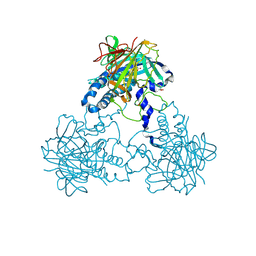 | | Crystal structure of human soluble Adenylyl Cyclase (Apo form) | | Descriptor: | ACETATE ION, ADENYLATE CYCLASE TYPE 10, GLYCEROL | | Authors: | Kleinboelting, S, Moniot, S, Weyand, M, Steegborn, C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-03-05 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Human Soluble Adenylyl Cyclase Reveal Mechanisms of Catalysis and of its Activation Through Bicarbonate.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BV2
 
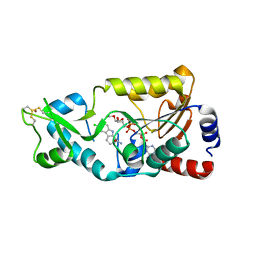 | | CRYSTAL STRUCTURE OF SIR2 IN COMPLEX WITH THE INHIBITOR EX-527, 2'-O-ACETYL-ADP-RIBOSE AND DEACETYLATED P53-PEPTIDE | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, CELLULAR TUMOR ANTIGEN P53, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2IYA
 
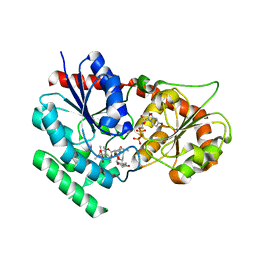 | | The crystal structure of macrolide glycosyltransferases: A blueprint for antibiotic engineering | | Descriptor: | (3S,5R,6S,7R,8R,11R,12S,13R,14S,15S)-6-HYDROXY-5,7,8,11,13,15-HEXAMETHYL-4,10-DIOXO-14-{[3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSYL]OXY}-1,9-DIOXASPIRO[2.13]HEXADEC-12-YL 2,6-DIDEOXY-3-O-METHYL-ALPHA-L-ARABINO-HEXOPYRANOSIDE, OLEANDOMYCIN GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Bolam, D.N, Roberts, S.M, Proctor, M.R, Turkenburg, J.P, Dodson, E.J, Martinez-Fleites, C, Yang, M, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2006-07-13 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Two Macrolide Glycosyltransferases Provides a Blueprint for Host Cell Antibiotic Immunity.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2IYF
 
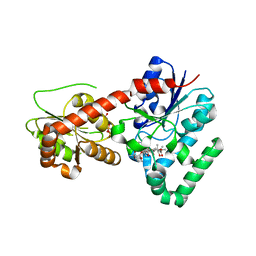 | | The crystal structure of macrolide glycosyltransferases: A blueprint for antibiotic engineering | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, OLEANDOMYCIN GLYCOSYLTRANSFERASE, ... | | Authors: | Bolam, D.N, Roberts, S.M, Proctor, M.R, Turkenburg, J.P, Dodson, E.J, Martinez-Fleites, C, Yang, M, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2006-07-17 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Two Macrolide Glycosyltransferases Provides a Blueprint for Host Cell Antibiotic Immunity.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
7BTH
 
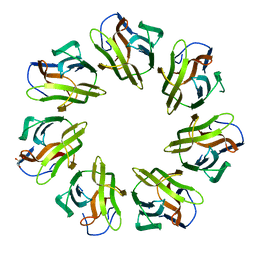 | | Mevo lectin- Native form-1 | | Descriptor: | GLYCEROL, lectin | | Authors: | Sivaji, N, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and related studies on Mevo lectin from Methanococcus voltae A3: the first thorough characterization of an archeal lectin and its interactions.
Glycobiology, 31, 2021
|
|
7BSN
 
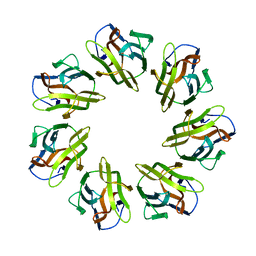 | | Mevo lectin complex with 3alpha-mannobiose | | Descriptor: | alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose, lectin | | Authors: | Sivaji, N, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2020-03-31 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and related studies on Mevo lectin from Methanococcus voltae A3: the first thorough characterization of an archeal lectin and its interactions.
Glycobiology, 31, 2021
|
|
7BT9
 
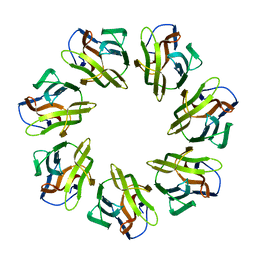 | | Mevo lectin complex with alpha-mannose | | Descriptor: | alpha-D-mannopyranose, lectin | | Authors: | Sivaji, N, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2020-03-31 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and related studies on Mevo lectin from Methanococcus voltae A3: the first thorough characterization of an archeal lectin and its interactions.
Glycobiology, 31, 2021
|
|
7BSB
 
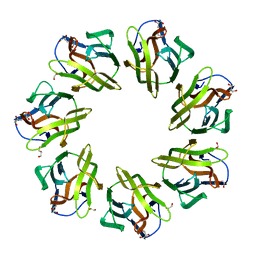 | | Mevo lectin- Native form-2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, lectin | | Authors: | Sivaji, N, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2020-03-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and related studies on Mevo lectin from Methanococcus voltae A3: the first thorough characterization of an archeal lectin and its interactions.
Glycobiology, 31, 2021
|
|
7BSM
 
 | | Mevo lectin complex with 2alpha-mannobiose | | Descriptor: | ALANINE, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Sivaji, N, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2020-03-31 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and related studies on Mevo lectin from Methanococcus voltae A3: the first thorough characterization of an archeal lectin and its interactions.
Glycobiology, 31, 2021
|
|
4BVG
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH NATIVE ALKYLIMIDATE FORMED FROM ACETYL-LYSINE ACS2-PEPTIDE CRYSTALLIZED IN PRESENCE OF THE INHIBITOR EX-527 | | Descriptor: | (2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL ACETATE, 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ex-527 inhibits Sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
1LJ3
 
 | |
1LJK
 
 | |
1LJ4
 
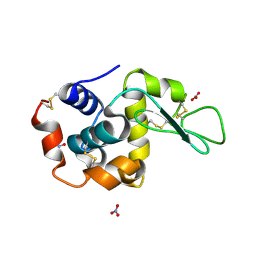 | |
1LJI
 
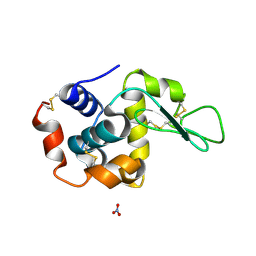 | |
1LJE
 
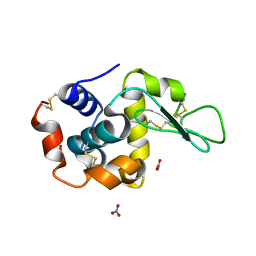 | |
4BN4
 
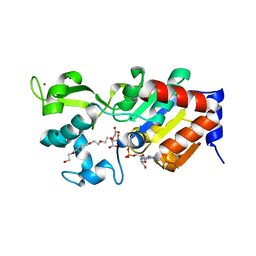 | | Structure of human SIRT3 in complex with ADP-ribose | | Descriptor: | 2-[2-[2-[2-[2-(2-hydroxyethyloxy)ethoxy]ethoxy]ethoxy]ethoxy]ethanoic acid, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-3, MITOCHONDRIAL, ... | | Authors: | Nguyen, G.T.T, Schaefer, S, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-05-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of Human Sirtuin 3 Complexes with Adp-Ribose and with Carba-Nad+ and Srt1720: Binding Details and Inhibition Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1LJJ
 
 | |
