6XNA
 
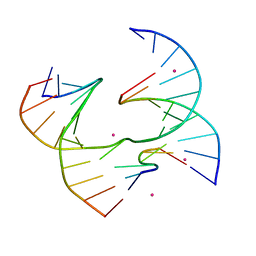 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J1 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*CP*TP*GP*AP*GP*TP*GP*TP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-02 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XO5
 
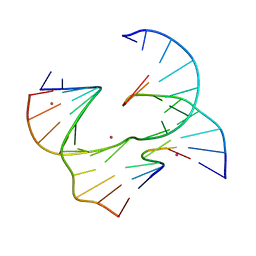 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J8 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-06 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.157 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XDZ
 
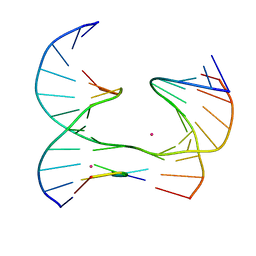 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J8 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-06-11 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.106 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XEK
 
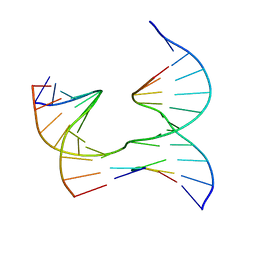 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J14 immobile Holliday junction | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-06-12 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XFD
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J20 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*CP*GP*TP*CP*TP*GP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-06-15 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.101 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XGJ
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J32 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*AP*GP*TP*CP*TP*GP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-06-17 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.071 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XFG
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J23 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-06-15 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.053 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XFW
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J24 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-06-16 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.053 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XO6
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J28 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*TP*GP*TP*CP*TP*GP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-06 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.108 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
4K12
 
 | | Structural Basis for Host Specificity of Factor H Binding by Streptococcus pneumoniae | | 分子名称: | Choline binding protein A, Complement factor H | | 著者 | Liu, A, Achila, D, Banerjee, R, Martinez-Hackert, E, Li, Y, Yan, H. | | 登録日 | 2013-04-04 | | 公開日 | 2014-04-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.079 Å) | | 主引用文献 | Structural determinants of host specificity of complement Factor H recruitment by Streptococcus pneumoniae.
Biochem.J., 465, 2015
|
|
4JPZ
 
 | | Voltage-gated sodium channel 1.2 C-terminal domain in complex with FGF13U and Ca2+/calmodulin | | 分子名称: | CALCIUM ION, Calmodulin, Fibroblast growth factor 13, ... | | 著者 | Wang, C, Chung, B.C, Yan, H, Wang, H.G, Lee, S.Y, Pitt, G.S. | | 登録日 | 2013-03-19 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | Structural analyses of Ca(2+)/CaM interaction with NaV channel C-termini reveal mechanisms of calcium-dependent regulation.
Nat Commun, 5, 2014
|
|
4JQ0
 
 | | Voltage-gated sodium channel 1.5 C-terminal domain in complex with FGF12B and Ca2+/calmodulin | | 分子名称: | CALCIUM ION, Calmodulin, Fibroblast growth factor 12, ... | | 著者 | Wang, C, Chung, B.C, Yan, H, Wang, H.G, Lee, S.Y, Pitt, G.S. | | 登録日 | 2013-03-19 | | 公開日 | 2014-05-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.84 Å) | | 主引用文献 | Structural analyses of Ca(2+)/CaM interaction with NaV channel C-termini reveal mechanisms of calcium-dependent regulation.
Nat Commun, 5, 2014
|
|
5FNU
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | (3S)-3-(7-methoxy-1-methyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)phenyl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2015-11-16 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNT
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | (3S)-3-{4-Chloro-3-[(N-methylbenzenesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2015-11-16 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FZJ
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | 2,6-DIMETHYL-4H-PYRANO[3,4-D][1,3]OXAZOL-4-ONE, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2016-03-14 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNR
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | (3S)-3-(4-chlorophenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2015-11-16 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNS
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | (3s)-{4-Chloro-3-[(N-methylmethanesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl) propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2015-11-16 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
2HK7
 
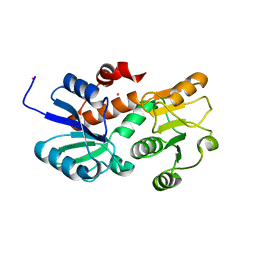 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus in complex with mercury at 2.5 angstrom resolution | | 分子名称: | MERCURY (II) ION, Shikimate dehydrogenase | | 著者 | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | 登録日 | 2006-07-03 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
2HK9
 
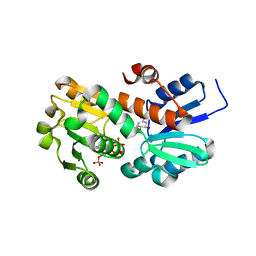 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus in complex with shikimate and NADP+ at 2.2 angstrom resolution | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | 登録日 | 2006-07-03 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
2HK8
 
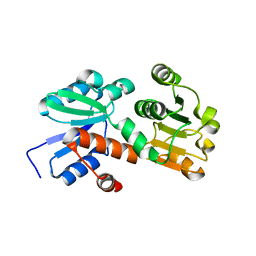 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus at 2.35 angstrom resolution | | 分子名称: | Shikimate dehydrogenase | | 著者 | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | 登録日 | 2006-07-03 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
2M6U
 
 | | NMR Structure of CbpAN from Streptococcus pneumoniae | | 分子名称: | Choline binding protein A | | 著者 | Liu, A, Yan, H, Achila, D, Martinez-Hackert, E, Li, Y, Banerjee, R. | | 登録日 | 2013-04-10 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural determinants of host specificity of complement Factor H recruitment by Streptococcus pneumoniae.
Biochem.J., 465, 2015
|
|
2O90
 
 | |
2NM2
 
 | |
2NM3
 
 | | Crystal structure of dihydroneopterin aldolase from S. aureus in complex with (1S,2S)-monapterin at 1.68 angstrom resolution | | 分子名称: | ACETATE ION, D-MONAPTERIN, Dihydroneopterin aldolase | | 著者 | Blaszczyk, J, Ji, X, Yan, H. | | 登録日 | 2006-10-20 | | 公開日 | 2007-09-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural basis for the aldolase and epimerase activities of Staphylococcus aureus dihydroneopterin aldolase.
J.Mol.Biol., 368, 2007
|
|
4US3
 
 | | Crystal Structure of the bacterial NSS member MhsT in an Occluded Inward-Facing State | | 分子名称: | DODECYL-ALPHA-D-MALTOSIDE, SODIUM ION, TRANSPORTER, ... | | 著者 | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | 登録日 | 2014-07-02 | | 公開日 | 2014-09-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.098 Å) | | 主引用文献 | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
