5Z0R
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
5Q02
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(1,2-oxazol-3-yl)thiophen-2-yl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-(1,2-oxazol-3-yl)thiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(1,2-oxazol-3-yl)thiophen-2-yl]sulfonylurea
To be published
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
1G0I
 
 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE-FRUCTOSE 1,6 BISPHOSPHATASE | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, INOSITOL MONOPHOSPHATASE, MANGANESE (II) ION, ... | | Authors: | Johnson, K.A, Chen, L, Yang, H, Roberts, M.F, Stec, B. | | Deposit date: | 2000-10-06 | | Release date: | 2001-03-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and catalytic mechanism of the MJ0109 gene product: a bifunctional enzyme with inositol monophosphatase and fructose 1,6-bisphosphatase activities.
Biochemistry, 40, 2001
|
|
5PZS
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(3-chlorophenyl)sulfonyl-3-[6-[(3-chlorophenyl)sulfonylcarbamoylamino]hexyl]urea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N,N'-(hexane-1,6-diyldicarbamoyl)bis(3-chlorobenzene-1-sulfonamide) | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-11 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(3-chlorophenyl)sulfonyl-3-[6-[(3-chlorophenyl)sulfonylcarbamoylamino]hexyl]urea
To be published
|
|
5Q06
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(2-methyl-1,3-thiazol-4-yl)phenyl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-4-(2-methyl-1,3-thiazol-4-yl)benzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(2-methyl-1,3-thiazol-4-yl)phenyl]sulfonylurea
To be published
|
|
5PZR
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(3-chlorophenyl)sulfonyl-3-[3-[3-[(3-chlorophenyl)sulfonylcarbamoylamino]propoxy]propyl]urea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N,N'-{oxybis[(propane-3,1-diyl)carbamoyl]}bis(3-chlorobenzene-1-sulfonamide) | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-11 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(3-chlorophenyl)sulfonyl-3-[3-[3-[(3-chlorophenyl)sulfonylcarbamoylamino]propoxy]propyl]urea
To be published
|
|
2JXB
 
 | | Structure of CD3epsilon-Nck2 first SH3 domain complex | | Descriptor: | T-cell surface glycoprotein CD3 epsilon chain, Cytoplasmic protein NCK2 | | Authors: | Takeuchi, K, Yang, H, Ng, E, Park, S, Sun, Z.J, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-11-09 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional evidence that Nck interaction with CD3epsilon regulates T-cell receptor activity.
J.Mol.Biol., 380, 2008
|
|
5PZV
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(4-bromopyridin-2-yl)-3-(4-chlorophenyl)sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(4-bromopyridin-2-yl)carbamoyl]-4-chlorobenzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-11 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(4-bromopyridin-2-yl)-3-(4-chlorophenyl)sulfonylurea
To be published
|
|
5Q0A
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-cyanopyrazin-2-yl)-3-[3-(difluoromethoxy)phenyl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-cyanopyrazin-2-yl)carbamoyl]-3-(difluoromethoxy)benzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-11 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-cyanopyrazin-2-yl)-3-[3-(difluoromethoxy)phenyl]sulfonylurea
To be published
|
|
1G0H
 
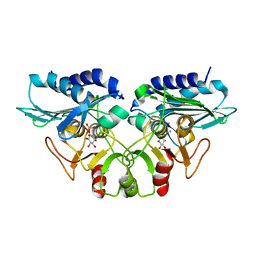 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE-FRUCTOSE 1,6 BISPHOSPHATASE | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1-PHOSPHATE, INOSITOL MONOPHOSPHATASE | | Authors: | Johnson, K.A, Chen, L, Yang, H, Roberts, M.F, Stec, B. | | Deposit date: | 2000-10-06 | | Release date: | 2001-03-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and catalytic mechanism of the MJ0109 gene product: a bifunctional enzyme with inositol monophosphatase and fructose 1,6-bisphosphatase activities.
Biochemistry, 40, 2001
|
|
5X8Y
 
 | | A Mutation identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in vitro | | Descriptor: | ZIKV NS1 | | Authors: | Wang, D, Chen, C, Liu, S, Zhou, H, Yang, K, Zhao, Q, Ji, X, Chen, C, Xie, W, Wang, Z, Mi, L.Z, Yang, H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Sci Rep, 7, 2017
|
|
1ZCK
 
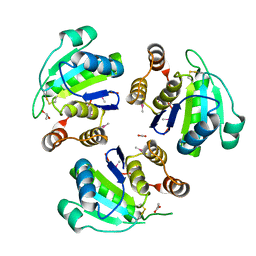 | | native structure prl-1 (ptp4a1) | | Descriptor: | ACETIC ACID, protein tyrosine phosphatase 4a1 | | Authors: | Sun, J.P, Wang, W.Q, Yang, H, Liu, S, Liang, F, Fedorov, A.A, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2005-04-12 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Biochemical Properties of PRL-1, a Phosphatase Implicated in Cell Growth, Differentiation, and Tumor Invasion.
Biochemistry, 44, 2005
|
|
4LTU
 
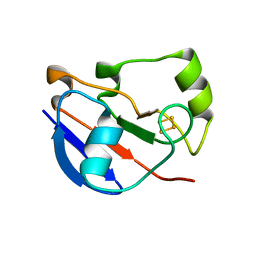 | | Crystal Structure of Ferredoxin from Rhodopseudomonas palustris HaA2 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Zhou, W.H, Zhang, T, Yang, H, Bell, S.G, Wong, L.-L. | | Deposit date: | 2013-07-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Ferredoxin from Rhodopseudomonas palustris HaA2
To be Published
|
|
2MNY
 
 | | NMR Structure of KDM5B PHD1 finger | | Descriptor: | Lysine-specific demethylase 5B, ZINC ION | | Authors: | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | Deposit date: | 2014-04-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
8UD2
 
 | | SARS-CoV-2 Nsp15, apo-form | | Descriptor: | Non-structural protein 15 | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 15, 2024
|
|
1X24
 
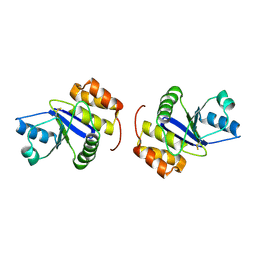 | | Prl-1 (ptp4a) | | Descriptor: | protein tyrosine phosphatase 4a1 | | Authors: | Zhang, Z.Y, Sun, J.P, Liu, S, Wang, W.Q, Yang, H. | | Deposit date: | 2005-04-20 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Biochemical Properties of PRL-1, a Phosphatase Implicated in Cell Growth, Differentiation, and Tumor Invasion(,)
Biochemistry, 44, 2005
|
|
1WP0
 
 | | Human SCO1 | | Descriptor: | SCO1 protein homolog | | Authors: | Williams, J.C, Sue, C, Banting, G.S, Yang, H, Glerum, D.M, Hendrickson, W.A, Schon, E.A. | | Deposit date: | 2004-08-27 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human SCO1: IMPLICATIONS FOR REDOX SIGNALING BY A MITOCHONDRIAL CYTOCHROME c OXIDASE "ASSEMBLY" PROTEIN
J.Biol.Chem., 280, 2005
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
8WDD
 
 | | Crystal structure of BSA in complex with B1 | | Descriptor: | Albumin, ~{N},~{N}-dimethyl-6-[(~{E})-2-(1-methylpyridin-1-ium-4-yl)ethenyl]naphthalen-2-amine | | Authors: | Chen, X, Ge, Y.H, Yang, H, Fang, B, Li, L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of BSA in complex with B1
To Be Published
|
|
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | Descriptor: | CidA_I gamma/2 protein, CidB_I b/2 protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | Descriptor: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIU
 
 | | Crystal structure of the DUB domain of Wolbachia cytoplasmic incompatibility factor CidB from wMel | | Descriptor: | ULP_PROTEASE domain-containing protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIT
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wMel | | Descriptor: | bacteria factor 1 | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
4GU0
 
 | | Crystal structure of LSD2 with H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Yang, H, Dong, Z, Fang, J, Zhu, T, Gong, W, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b
Cell Res., 23, 2013
|
|
