7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp15/16 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp9/10 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp6/7 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4FTX
 
 | | Crystal structure of Ego3 homodimer | | 分子名称: | Protein SLM4, SUCCINIC ACID | | 著者 | Zhang, T, Peli-Gulli, M.P, Yang, H, De Virgilio, C, Ding, J. | | 登録日 | 2012-06-28 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1.
Structure, 20, 2012
|
|
1G0I
 
 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE-FRUCTOSE 1,6 BISPHOSPHATASE | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, INOSITOL MONOPHOSPHATASE, MANGANESE (II) ION, ... | | 著者 | Johnson, K.A, Chen, L, Yang, H, Roberts, M.F, Stec, B. | | 登録日 | 2000-10-06 | | 公開日 | 2001-03-14 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure and catalytic mechanism of the MJ0109 gene product: a bifunctional enzyme with inositol monophosphatase and fructose 1,6-bisphosphatase activities.
Biochemistry, 40, 2001
|
|
3GFC
 
 | | Crystal Structure of Histone-binding protein RBBP4 | | 分子名称: | Histone-binding protein RBBP4 | | 著者 | Amaya, M.F, Dong, A, Li, Z, He, H, Ni, S, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | 登録日 | 2009-02-26 | | 公開日 | 2009-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
1ZCL
 
 | | prl-1 c104s mutant in complex with sulfate | | 分子名称: | SULFATE ION, protein tyrosine phosphatase 4a1 | | 著者 | Sun, J.P, Wang, W.Q, Yang, H, Liu, S, Liang, F, Fedorov, A.A, Almo, S.C, Zhang, Z.Y. | | 登録日 | 2005-04-12 | | 公開日 | 2005-09-20 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure and Biochemical Properties of PRL-1, a Phosphatase Implicated in Cell Growth, Differentiation, and Tumor Invasion.
Biochemistry, 44, 2005
|
|
8H68
 
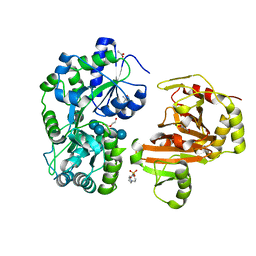 | | Crystal structure of Caenorhabditis elegans NMAD-1 in complex with NOG and Mg(II) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Shi, Y, Ding, J, Yang, H. | | 登録日 | 2022-10-16 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Caenorhabditis elegans NMAD-1 functions as a demethylase for actin.
J Mol Cell Biol, 15, 2023
|
|
7DW2
 
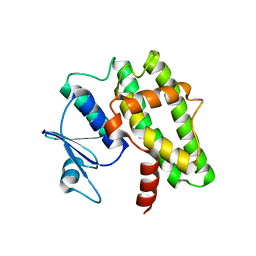 | |
5F5A
 
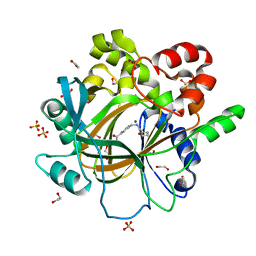 | | Crystal Structure of human JMJD2D complexed with KDOAM16 | | 分子名称: | 1,2-ETHANEDIOL, 2-[(furan-2-ylmethylamino)methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4D, ... | | 著者 | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-12-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
8HWN
 
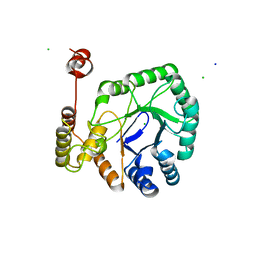 | |
5F5I
 
 | | Crystal Structure of human JMJD2A complexed with KDOOA011340 | | 分子名称: | 2-[[(phenylmethyl)amino]methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | 著者 | Krojer, T, Vollmar, M, Crawley, L, Szykowska, A, Gileadi, C, Johansson, C, England, K, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-12-04 | | 公開日 | 2015-12-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5D47
 
 | | Crystal Structure of FABP4 in complex with 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl] propanoic acid | | 分子名称: | 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl]propanoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | 登録日 | 2015-08-07 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
6BWJ
 
 | | Crystal structure of the TRPV2 ion channel in complex with RTx | | 分子名称: | CALCIUM ION, Transient receptor potential cation channel subfamily V member 2, resiniferatoxin | | 著者 | Zubcevic, L, Le, S, Yang, H, Lee, S.Y. | | 登録日 | 2017-12-15 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Conformational plasticity in the selectivity filter of the TRPV2 ion channel.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7E58
 
 | | interferon-inducible anti-viral protein 2 | | 分子名称: | Guanylate-binding protein 2 | | 著者 | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-02-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E59
 
 | | interferon-inducible anti-viral protein truncated | | 分子名称: | Guanylate-binding protein 5 | | 著者 | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-02-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5WVR
 
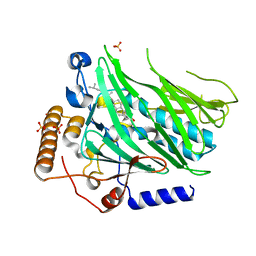 | | Crystal structure of Osh1 ORD domain in complex with cholesterol | | 分子名称: | CHOLESTEROL, KLLA0C04147p, SULFATE ION | | 著者 | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | 登録日 | 2016-12-28 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
6BWM
 
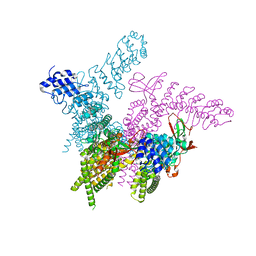 | | Crystal structure of the TRPV2 ion channel | | 分子名称: | CALCIUM ION, Transient receptor potential cation channel subfamily V member 2 | | 著者 | Zubcevic, L, Le, S, Yang, H, Lee, S.Y. | | 登録日 | 2017-12-15 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Conformational plasticity in the selectivity filter of the TRPV2 ion channel.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8UD2
 
 | | SARS-CoV-2 Nsp15, apo-form | | 分子名称: | Non-structural protein 15 | | 著者 | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | 登録日 | 2023-09-28 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.33 Å) | | 主引用文献 | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 15, 2024
|
|
5W45
 
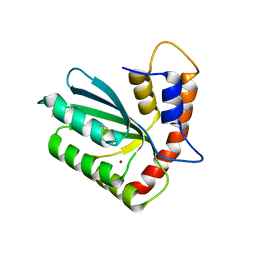 | | Crystal structure of APOBEC3H | | 分子名称: | APOBEC3H, ZINC ION | | 著者 | Ito, F, Yang, H.J, Xiao, X, Li, S.X, Wolfe, A, Zirkle, B, Arutiunian, V, Chen, X.S. | | 登録日 | 2017-06-09 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.486 Å) | | 主引用文献 | Understanding the Structure, Multimerization, Subcellular Localization and mC Selectivity of a Genomic Mutator and Anti-HIV Factor APOBEC3H.
Sci Rep, 8, 2018
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | 分子名称: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | 登録日 | 2015-08-07 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
1DK4
 
 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE | | 分子名称: | INOSITOL MONOPHOSPHATASE, PHOSPHATE ION, ZINC ION | | 著者 | Stec, B, Yang, H, Johnson, K.A, Chen, L, Roberts, M.F. | | 登録日 | 1999-12-06 | | 公開日 | 2000-11-08 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | MJ0109 is an enzyme that is both an inositol monophosphatase and the 'missing' archaeal fructose-1,6-bisphosphatase.
Nat.Struct.Biol., 7, 2000
|
|
8U78
 
 | | Structure of a N-Me-D-Gln4,Lys10-teixobactin analogue | | 分子名称: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, N-Methyl-D-Gln4,Lys10-teixobactin | | 著者 | Nowick, J.S, Yang, H, Kreutzer, A.G. | | 登録日 | 2023-09-14 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Supramolecular Interactions of Teixobactin Analogues in the Crystal State.
J.Org.Chem., 89, 2024
|
|
3TZD
 
 | | Crystal structure of the complex of Human Chromobox Homolog 3 (CBX3) | | 分子名称: | Chromobox protein homolog 3, Histone H1.4 | | 著者 | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | 登録日 | 2011-09-27 | | 公開日 | 2012-03-07 | | 最終更新日 | 2013-01-23 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
