8PUG
 
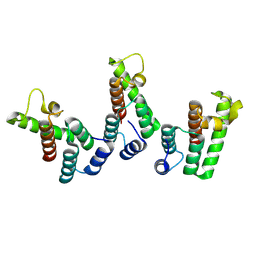 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement | | 分子名称: | Gag polyprotein | | 著者 | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | 登録日 | 2023-07-17 | | 公開日 | 2023-08-23 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
2RJE
 
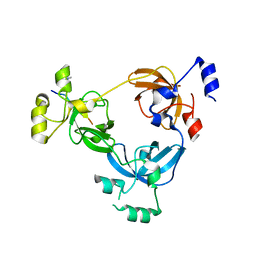 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 17-25), orthorhombic form II | | 分子名称: | CHLORIDE ION, Histone H4, Lethal(3)malignant brain tumor-like protein | | 著者 | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | 登録日 | 2007-10-14 | | 公開日 | 2007-10-30 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3MQ9
 
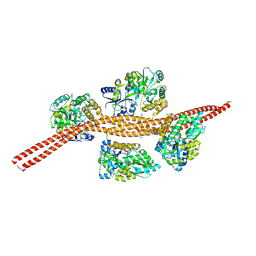 | |
5QD7
 
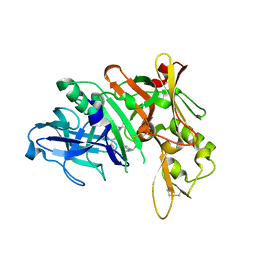 | | Crystal structure of BACE complex with BMC014 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({1-[3-(1-methylethyl)phenyl]cyclopropyl}amino)ethyl]-19-(methoxymethyl)-11-oxa-3,16-diazatric yclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
3CMT
 
 | | Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*DTP*DTP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DCP*DTP*DTP*DTP*DT)-3'), DNA (5'-D(P*DGP*DGP*DTP*DGP*DGP*DG)-3'), ... | | 著者 | Chen, Z, Yang, H, Pavletich, N.P. | | 登録日 | 2008-03-24 | | 公開日 | 2008-05-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures.
Nature, 453, 2008
|
|
6PSL
 
 | | Structure of a N-Me-D-Gln4,D-aza-Thr8,Arg10-teixobactin analogue | | 分子名称: | CHLORIDE ION, teixobactin analogue | | 著者 | Nowick, J.S, Yang, H, Pishenko, A, Li, X. | | 登録日 | 2019-07-12 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Design, Synthesis, and Study of Lactam and Ring-Expanded Analogues of Teixobactin.
J.Org.Chem., 85, 2020
|
|
8GX3
 
 | | The crystal structure of human Calpain-1 protease core in complex with 14c | | 分子名称: | CALCIUM ION, Calpain-1 catalytic subunit, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | 著者 | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | 登録日 | 2022-09-18 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | The crystal structure of human Calpain-1 protease core in complex with 14c
To Be Published
|
|
8GXG
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14a | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | 著者 | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | 登録日 | 2022-09-20 | | 公開日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | The crystal structure of SARS-CoV-2 main protease in complex with 14a
To Be Published
|
|
8GXH
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14b | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | 著者 | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | 登録日 | 2022-09-20 | | 公開日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | The crystal structure of SARS-CoV-2 main protease in complex with 14b
To Be Published
|
|
5Q1H
 
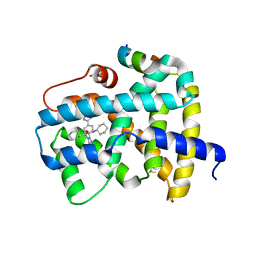 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-N,2-dicyclohexyl-2-{2-[4-(1H-tetrazol-5-yl)phenyl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5GJB
 
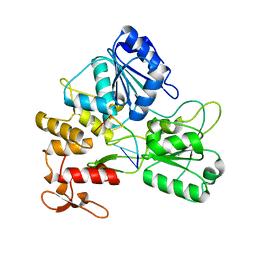 | | Zika virus NS3 helicase in complex with ssRNA | | 分子名称: | NS3 helicase, RNA (5'-R(*AP*GP*AP*UP*CP*AP*A)-3') | | 著者 | Tian, H.L, Ji, X.Y, Yang, X.Y, Zhang, Z.X, Lu, Z.K, Yang, K.L, Chen, C, Zhao, Q, Chi, H, Mu, Z.Y, Xie, W, Wang, Z.F, Lou, H.Q, Yang, H.T, Rao, Z.H. | | 登録日 | 2016-06-28 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.702 Å) | | 主引用文献 | Structural basis of Zika virus helicase in recognizing its substrates
Protein Cell, 7, 2016
|
|
2RJD
 
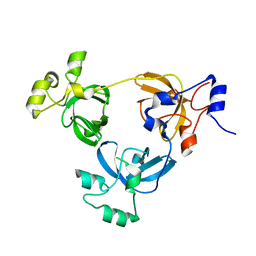 | | Crystal structure of L3MBTL1 protein | | 分子名称: | Lethal(3)malignant brain tumor-like protein | | 著者 | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | 登録日 | 2007-10-14 | | 公開日 | 2007-10-30 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | 登録日 | 2020-01-26 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
5KK5
 
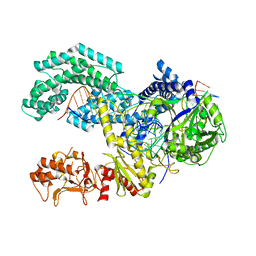 | | AsCpf1(E993A)-crRNA-DNA ternary complex | | 分子名称: | CRISPR-associated endonuclease Cpf1, DNA (28-MER), DNA (8-mer), ... | | 著者 | Gao, P, Yang, H, Rajashankar, K.R, Huang, Z, Patel, D.J. | | 登録日 | 2016-06-21 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.289 Å) | | 主引用文献 | Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition.
Cell Res., 26, 2016
|
|
5Q0O
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-{2-[4-(benzenecarbonyl)phenyl]-1H-benzimidazol-1-yl}-N,2-dicyclohexylacetamide, Bile acid receptor, CHLORIDE ION, ... | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q14
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 4-{(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylethoxy}-3-fluorobenzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0U
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, trans-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)cyclohexyl hydrogen sulfate | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1C
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-cyclohexyl-2-[2-(2,6-dimethoxypyridin-3-yl)-5,6-difluoro-1H-benzimidazol-1-yl]-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0N
 
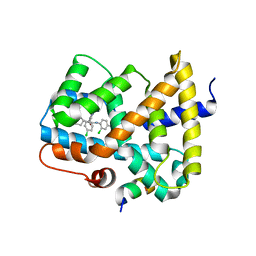 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 3-chloro-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)benzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1D
 
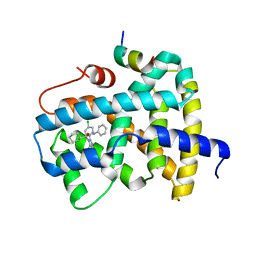 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexyl-N-phenylacetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0J
 
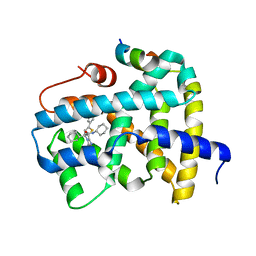 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-N,2-dicyclohexyl-2-[2-(5-phenylthiophen-2-yl)-1H-benzimidazol-1-yl]acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q18
 
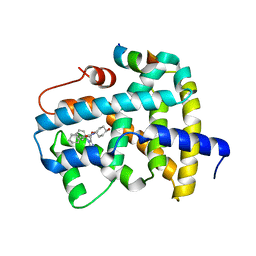 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-cyclohexyl-2-{5,6-difluoro-2-[(R)-methoxy(phenyl)methyl]-1H-benzimidazol-1-yl}-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q15
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-N,2-dicyclohexyl-2-{5,6-difluoro-2-[(R)-methoxy(phenyl)methyl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0P
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 4-{(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylethoxy}benzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q16
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-{2-[(4-chloro-2-methylphenoxy)methyl]-6-fluoro-1H-benzimidazol-1-yl}-N,2-dicyclohexylacetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
