3WT5
 
 | | A mixed population of antagonist and agonist binding conformers in a single crystal explains partial agonism against vitamin D receptor: Active vitamin D analogues with 22R-alkyl group | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3R)-3-butyl-6-hydroxy-6-methylheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Mixed Population of Antagonist and Agonist Binding Conformers in a Single Crystal Explains Partial Agonism against Vitamin D Receptor: Active Vitamin D Analogues with 22R-Alkyl Group.
J.Med.Chem., 57, 2014
|
|
3WTQ
 
 | | Crystal structure of VDR-LBD complexed with 22S-butyl-2-methylidene-19-nor-1a,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3S)-3-butyl-6-hydroxy-6-methylheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Inaba, Y, Itoh, T, Anami, Y, Ikura, T, Ito, N, Yamamoto, K. | | Deposit date: | 2014-04-15 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of VDR-LBD complexed with 22S-butyl-2-methylidene-19-nor-1a,25-dihydroxyvitamin D3
To be Published
|
|
3WBP
 
 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-1) | | Descriptor: | 1,2-ETHANEDIOL, C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WBR
 
 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-3) | | Descriptor: | C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WT7
 
 | | Crystal structure of VDR-LBD complexed with 22R-Butyl-2-methylidene-26,27-dimethyl-19,24-dinor-1 ,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3R)-3-butyl-5-ethyl-5-hydroxyheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Mixed Population of Antagonist and Agonist Binding Conformers in a Single Crystal Explains Partial Agonism against Vitamin D Receptor: Active Vitamin D Analogues with 22R-Alkyl Group.
J.Med.Chem., 57, 2014
|
|
3WT6
 
 | | A mixed population of antagonist and agonist binding conformers in a single crystal explains partial agonism against vitamin D receptor: Active vitamin D analogues with 22R-alkyl group | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3R)-3-butyl-6-hydroxy-6-methylheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mixed Population of Antagonist and Agonist Binding Conformers in a Single Crystal Explains Partial Agonism against Vitamin D Receptor: Active Vitamin D Analogues with 22R-Alkyl Group.
J.Med.Chem., 57, 2014
|
|
3WC6
 
 | | Carboxypeptidase B in complex with 2nd zinc | | Descriptor: | ACETATE ION, Carboxypeptidase B, ZINC ION | | Authors: | Yoshimoto, N, Itoh, T, Inaba, Y, Yamamoto, K. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for inhibition of carboxypeptidase B by selenium-containing inhibitor: selenium coordinates to zinc in enzyme.
J.Med.Chem., 56, 2013
|
|
3X1H
 
 | | hPPARgamma Ligand binding domain in complex with 5-oxo-tricosahexaenoic acid | | Descriptor: | (7E,11Z,14Z,17Z,20Z)-5-oxotricosa-7,11,14,17,20-pentaenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-11-18 | | Release date: | 2015-04-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of covalent bond formation between PPAR gamma and oxo-fatty acids.
Bioconjug.Chem., 26, 2015
|
|
3X1I
 
 | | hPPARgamma Ligand binding domain in complex with 6-oxo-tetracosahexaenoic acid | | Descriptor: | (8E,12Z,15Z,18Z,21Z)-6-oxotetracosa-8,12,15,18,21-pentaenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-11-18 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of covalent bond formation between PPAR gamma and oxo-fatty acids.
Bioconjug.Chem., 26, 2015
|
|
7EFL
 
 | | Crystal structure of the gastric proton pump K791S in (BYK)E2BeF state | | Descriptor: | (7R,8R,9S)-2,3-dimethyl-9-phenyl-7,8,9,10-tetrahydroimidazo[1,2-h][1,7]naphthyridine-7,8-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Abe, K, Yamamoto, K, Irie, K. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Gastric proton pump with two occluded K + engineered with sodium pump-mimetic mutations.
Nat Commun, 12, 2021
|
|
7EFM
 
 | | Crystal structure of the gastric proton pump K791S/E820D/Y340N in (BYK)E2BeF state | | Descriptor: | (7R,8R,9S)-2,3-dimethyl-9-phenyl-7,8,9,10-tetrahydroimidazo[1,2-h][1,7]naphthyridine-7,8-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Abe, K, Yamamoto, K, Irie, K. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Gastric proton pump with two occluded K + engineered with sodium pump-mimetic mutations.
Nat Commun, 12, 2021
|
|
7EFN
 
 | | Crystal structure of the gastric proton pump K791S/E820D/Y340N/E936V in (BYK)E2BeF state | | Descriptor: | (7R,8R,9S)-2,3-dimethyl-9-phenyl-7,8,9,10-tetrahydroimidazo[1,2-h][1,7]naphthyridine-7,8-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Abe, K, Yamamoto, K, Irie, K. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Gastric proton pump with two occluded K + engineered with sodium pump-mimetic mutations.
Nat Commun, 12, 2021
|
|
6JXK
 
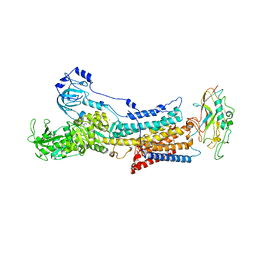 | | Rb+-bound E2-MgF state of the gastric proton pump (Wild-type) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Abe, K, Irie, K, Yamamoto, K. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | A single K + -binding site in the crystal structure of the gastric proton pump.
Elife, 8, 2019
|
|
6K0T
 
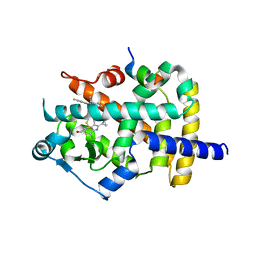 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-17 | | Descriptor: | 3-[(1~{E})-1-[8-[(8-chloranyl-2-cyclopropyl-imidazo[1,2-a]pyridin-3-yl)methyl]-3-fluoranyl-6~{H}-benzo[c][1]benzoxepin-11-ylidene]ethyl]-4~{H}-1,2,4-oxadiazol-5-one, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Suzuki, M, Yamamoto, K, Takahashi, Y, Saito, J. | | Deposit date: | 2019-05-07 | | Release date: | 2019-10-30 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Development of a novel class of peroxisome proliferator-activated receptor (PPAR) gamma ligands as an anticancer agent with a unique binding mode based on a non-thiazolidinedione scaffold.
Bioorg.Med.Chem., 27, 2019
|
|
6J7A
 
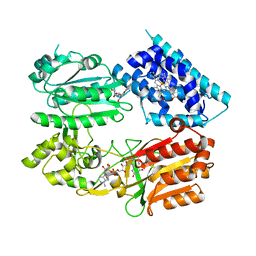 | | Fusion protein of heme oxygenase-1 and NADPH cytochrome P450 reductase (17aa) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1,NADPH--cytochrome P450 reductase, ... | | Authors: | Sugishima, M, Sato, H, Wada, K, Yamamoto, K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.269 Å) | | Cite: | Crystal structure of a NADPH-cytochrome P450 oxidoreductase (CYPOR) and heme oxygenase 1 fusion protein implies a conformational change in CYPOR upon NADPH/NADP+binding.
Febs Lett., 593, 2019
|
|
6J7I
 
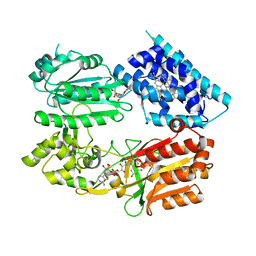 | | Fusion protein of heme oxygenase-1 and NADPH cytochrome P450 reductase (15aa) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1,NADPH--cytochrome P450 reductase, ... | | Authors: | Sugishima, M, Sato, H, Wada, K, Yamamoto, K. | | Deposit date: | 2019-01-18 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a NADPH-cytochrome P450 oxidoreductase (CYPOR) and heme oxygenase 1 fusion protein implies a conformational change in CYPOR upon NADPH/NADP+binding.
Febs Lett., 593, 2019
|
|
2LQI
 
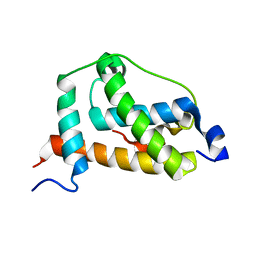 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2l3b conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LQH
 
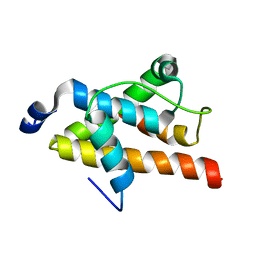 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2M33
 
 | | Solution NMR structure of full-length oxidized microsomal rabbit cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Subramanian, V, Ahuja, S, Popovych, N, Huang, R, Le Clair, S.V, Jahr, N, Soong, R, Xu, J, Yamamoto, K, Nanga, R.P, Im, S, Waskell, L, Ramamoorthy, A. | | Deposit date: | 2013-01-08 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of full-length mammalian cytochrome b5
To be Published
|
|
6JEY
 
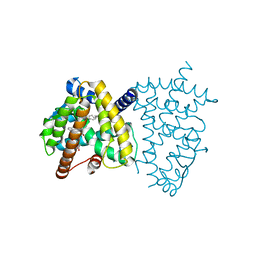 | | Covalent bond formation between ynone moiety of synthetic fatty acid and hPPARg-LBD | | Descriptor: | (9Z,12Z,15Z,18Z,21Z)-5-oxidanylidenetetracosa-9,12,15,18,21-pentaen-6-ynoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Yamamoto, K, Itoh, T. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cyclization Reaction-Based Turn-on Probe for Covalent Labeling of Target Proteins.
Cell Chem Biol, 27, 2020
|
|
6JEZ
 
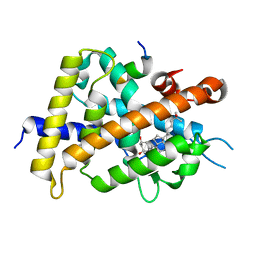 | | Covalent labeling of rVDR-LBD by turn-on fluorescent probe mediated by conjugate addition and cyclization | | Descriptor: | (1R,3R)-5-(2-((1R,3aS,7aR,E)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methyloctahydro-4H-inden-4-ylidene)ethylidene)-2- methylenecyclohexane-1,3-diol, 7-(diethylamino)chromen-2-one, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kojima, H, Yamamoto, K, Itoh, T. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cyclization Reaction-Based Turn-on Probe for Covalent Labeling of Target Proteins.
Cell Chem Biol, 27, 2020
|
|
6JF0
 
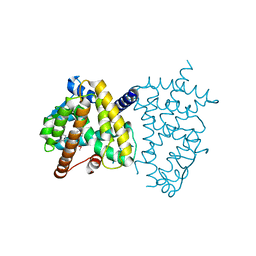 | |
6IQ5
 
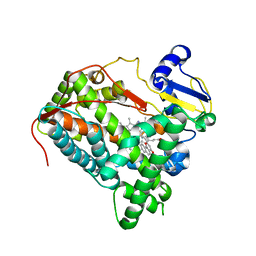 | | Crystal Structure of CYP1B1 and Inhibitor Having Azide Group | | Descriptor: | 2-(cis-4-azidocyclohexyl)-4H-naphtho[1,2-b]pyran-4-one, Cytochrome P450 1B1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kubo, M, Yamamoto, K, Itoh, T. | | Deposit date: | 2018-11-06 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design and synthesis of selective CYP1B1 inhibitor via dearomatization of alpha-naphthoflavone.
Bioorg. Med. Chem., 27, 2019
|
|
5AZT
 
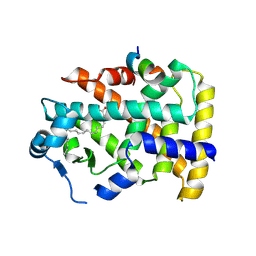 | | Ternary complex of hPPARalpha ligand binding domain, 17-oxoDHA and a SRC1 peptide | | Descriptor: | (4~{Z},7~{Z},10~{Z},13~{Z},19~{Z})-17-oxidanylidenedocosa-4,7,10,13,19-pentaenoic acid, 15-meric peptide from Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2015-10-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | 17-OxoDHA Is a PPAR alpha/gamma Dual Covalent Modifier and Agonist
Acs Chem.Biol., 2016
|
|
5AJ0
 
 | | Cryo electron microscopy of actively translating human polysomes (POST state). | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Behrmann, E, Loerke, J, Budkevich, T.V, Yamamoto, K, Schmidt, A, Penczek, P.A, Vos, M.R, Burger, J, Mielke, T, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-02-19 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Snapshots of Actively Translating Human Ribosomes
Cell(Cambridge,Mass.), 161, 2015
|
|
