3A4U
 
 | | Crystal structure of MCFD2 in complex with carbohydrate recognition domain of ERGIC-53 | | Descriptor: | CALCIUM ION, GLYCEROL, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Nishio, M, Kamiya, Y, Mizushima, T, Wakatsuki, S, Sasakawa, H, Yamamoto, K, Uchiyama, S, Noda, M, McKay, A.R, Fukui, K, Hauri, H.P, Kato, K. | | Deposit date: | 2009-07-17 | | Release date: | 2010-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the cooperative interplay between the two causative gene products of combined factor V and factor VIII deficiency.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5ZF1
 
 | | Molecular structure of a novel 5,10-methylenetetrahydrofolate dehydrogenase from the silkworm, Bombyx mori | | Descriptor: | 1,2-ETHANEDIOL, 5,10-methylenetetrahydrofolate dehydrogenase, SULFATE ION | | Authors: | Haque, R, Higashiura, A, Nakagawa, A, Yamamoto, K. | | Deposit date: | 2018-03-02 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular structure of a 5,10-methylenetetrahydrofolate dehydrogenase from the silkwormBombyx mori.
FEBS Open Bio, 9, 2019
|
|
5ZEQ
 
 | | Carboxypeptidase B in complex with DD28 | | Descriptor: | (2~{S})-2-[(6-azanyl-5-chloranyl-pyridin-3-yl)methyl]-3-selanyl-propanoic acid, CACODYLATE ION, Carboxypeptidase B, ... | | Authors: | Itoh, T, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the selective inhibition of activated thrombin-activatable fibrinolysis inhibitor (TAFIa) by a selenium-containing inhibitor with chloro-aminopyridine as a basic group
Bioorg. Med. Chem. Lett., 28, 2018
|
|
3AY8
 
 | | Glutathione S-transferase unclassified 2 from Bombyx mori | | Descriptor: | GLYCEROL, Glutathione S-transferase | | Authors: | Kakuta, Y, Usuda, K, Nakashima, T, Kimura, M, Aso, Y, Yamamoto, K. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic survey of active sites of an unclassified glutathione transferase from Bombyx mori
Biochim.Biophys.Acta, 1810, 2011
|
|
3G8X
 
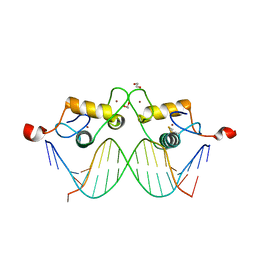 | | GR DNA binding domain:GilZ 16bp complex-65 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*TP*TP*GP*GP*GP*TP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*CP*CP*CP*AP*AP*TP*GP*TP*TP*CP*T)-3'), ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-12 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G9J
 
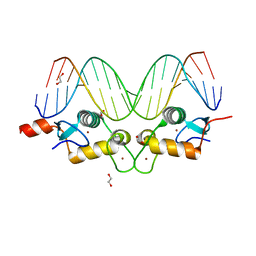 | | GR DNA binding domain:Pal, 18bp complex-36 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*AP*AP*AP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G9M
 
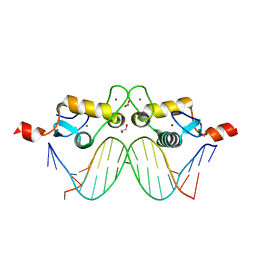 | | GR DNA-binding domain:Sgk 16bp complex-44 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*CP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*GP*AP*CP*AP*AP*AP*AP*TP*GP*TP*TP*CP*T)-3'), ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G6P
 
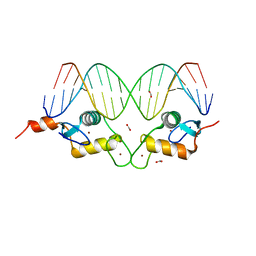 | | GR DNA binding domain:FKBP5 complex, 18bp | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*CP*CP*CP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*GP*GP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-07 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G6R
 
 | | GR DNA binding domain:FKBP5 complex-52, 18bp | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*CP*CP*CP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*GP*GP*TP*GP*TP*TP*CP*TP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-08 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G6T
 
 | | GR gamma DNA-binding domain:FKBP5 16bp complex-34 | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*GP*GP*GP*TP*GP*TP*TP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*AP*AP*CP*AP*CP*CP*CP*TP*GP*TP*TP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-08 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G6Q
 
 | | GR DNA binding domain:FKBP5 binding site complex-9 | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*CP*CP*CP*TP*GP*TP*TP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*AP*AP*CP*AP*GP*GP*GP*TP*GP*TP*TP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-08 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G99
 
 | | GR DNA binding domain:Pal complex-9 | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*TP*CP*T)-3'), ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G9P
 
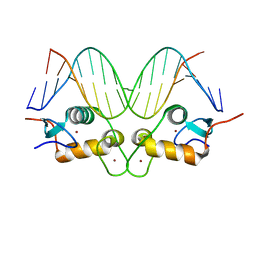 | | GR DNA binding domain:Sgk 16bp complex-7 | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*CP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*GP*AP*CP*AP*AP*AP*AP*TP*GP*TP*TP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G9I
 
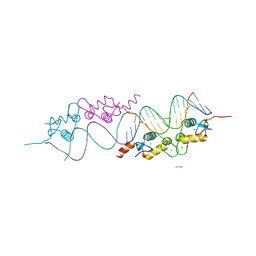 | | GR DNA Binding domain: Pal complex-35 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*TP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*AP*AP*CP*AP*AP*AP*AP*TP*GP*TP*TP*CP*T)-3'), ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G9O
 
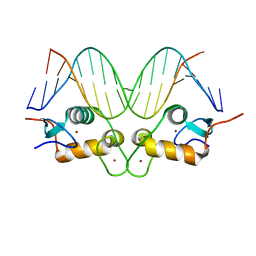 | | GR DNA-binding domain:Sgk 16bp complex-9 | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*CP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*GP*AP*CP*AP*AP*AP*AP*TP*GP*TP*TP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G97
 
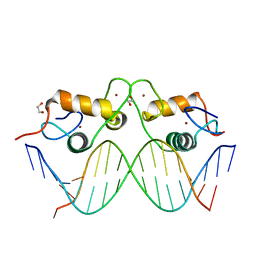 | | GR DNA-binding domain:GilZ 16bp complex-9 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*TP*TP*GP*GP*GP*TP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*CP*CP*CP*AP*AP*TP*GP*TP*TP*CP*T)-3'), ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
5XPL
 
 | | Crystal structure of VDR-LBD complexed with 22S-butyl-25-hydroxyphenyl-2-methylidene-19,26,27-trinor-25-oxo-1-hydroxyvitamin D3 | | Descriptor: | (4~{S})-4-[(1~{R})-1-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl )cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]ethyl]-1-(4-hydroxyphenyl)octan-1-one, Nuclear receptor coactivator 2, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
3VK9
 
 | | Crystal structure of delta-class glutathione transferase from silkmoth | | Descriptor: | GLYCEROL, Glutathione S-transferase delta | | Authors: | Kakuta, Y, Usuda, K, Higashiura, A, Suzuki, M, Nakagawa, A, Kimura, M, Yamamoto, K. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for catalytic activity of a silkworm Delta-class glutathione transferase
Biochim.Biophys.Acta, 1820, 2012
|
|
3VYJ
 
 | | Crystal structure of C-type lectin domain of murine dendritic cell inhibitory receptor 2 (apo form) | | Descriptor: | C-type lectin domain family 4, member a4, SULFATE ION | | Authors: | Nagae, M, Yamanaka, K, Hanashima, S, Ikeda, A, Satoh, T, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2012-09-26 | | Release date: | 2013-10-02 | | Last modified: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Recognition of Bisecting N-Acetylglucosamine: STRUCTURAL BASIS FOR ASYMMETRIC INTERACTION WITH THE MOUSE LECTIN DENDRITIC CELL INHIBITORY RECEPTOR 2
J.Biol.Chem., 288, 2013
|
|
5ZWI
 
 | | Interaction between Vitamin D receptor (VDR) and a ligand having a dienone group | | Descriptor: | (2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-4,6-diene-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
5ZWH
 
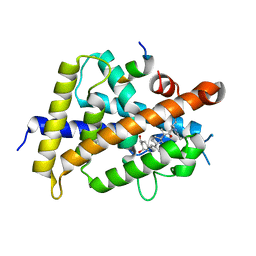 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having an ene-ynone group via conjugate addition reaction | | Descriptor: | (2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-4,6-diene-3-one, (E,2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-6-en-4-yn-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, ... | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
5XPM
 
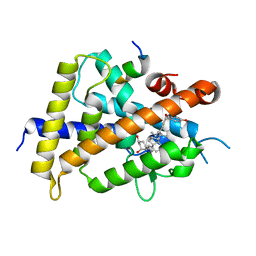 | | Crystal structure of VDR-LBD complexed with 22S-Butyl-25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},3~{S})-3-[(3~{S})-3-(4-hydroxyphenyl)-3-methoxy-propyl]heptan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5XPN
 
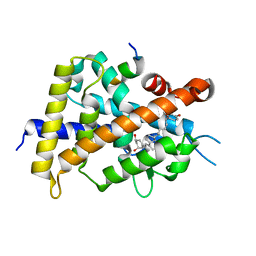 | | Crystal structure of VDR-LBD complexed with 25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{S})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5XPP
 
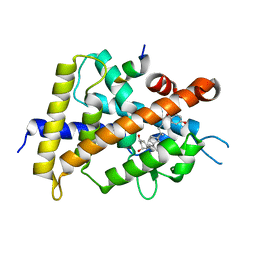 | | Crystal structure of VDR-LBD complexed with 25RS-(Hydroxyphenyl)-2-methylidene-19,26,27-trinor-1,25-dihydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-oxidanyl-hexan-2-yl]-7~{ a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5ZWE
 
 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having a vinyl ketone group via conjugate addition reaction | | Descriptor: | (6R)-6-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]hept-1-en-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
