2BQG
 
 | |
2BQD
 
 | |
2BQL
 
 | |
6IJY
 
 | |
7EF8
 
 | | Crystal structure of mouse MUTYH in complex with DNA containing AP site analogue:8-oxoG (Form I) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*TP*AP*GP*TP*CP*CP*CP*(3DR)P*GP*TP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*CP*T)-3'), ... | | Authors: | Nakamura, T, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2021-03-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the mammalian adenine DNA glycosylase MUTYH: insights into the base excision repair pathway and cancer.
Nucleic Acids Res., 49, 2021
|
|
7EFA
 
 | |
7EF9
 
 | | Crystal structure of mouse MUTYH in complex with DNA containing AP site analogue:8-oxoG (Form II) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*AP*TP*GP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*CP*CP*CP*(3DR)P*GP*TP*CP*TP*C)-3'), ... | | Authors: | Nakamura, T, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2021-03-21 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the mammalian adenine DNA glycosylase MUTYH: insights into the base excision repair pathway and cancer.
Nucleic Acids Res., 49, 2021
|
|
6L9W
 
 | | Crystal structure of mouse TIFA (T9E/C36S mutant) | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
6L9V
 
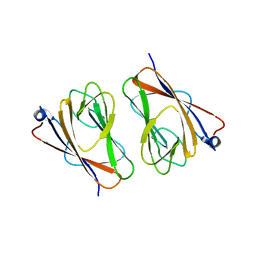 | | Crystal structure of mouse TIFA (T9D/C36S mutant) | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
1C7P
 
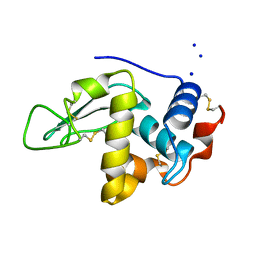 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME WITH FOUR EXTRA RESIDUES (EAEA) AT THE N-TERMINAL | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Goda, S, Takano, K, Yamagata, Y, Katakura, Y, Yutani, K. | | Deposit date: | 2000-02-29 | | Release date: | 2000-04-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effect of extra N-terminal residues on the stability and folding of human lysozyme expressed in Pichia pastoris.
Protein Eng., 13, 2000
|
|
6L9U
 
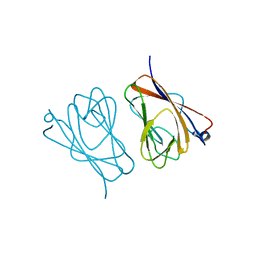 | | Crystal structure of mouse TIFA | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
1ISE
 
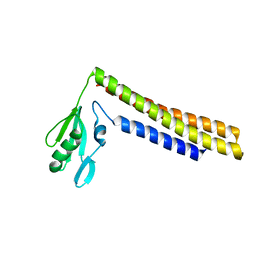 | | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly | | Descriptor: | Ribosome Recycling Factor | | Authors: | Nakano, H, Yoshida, T, Oka, S, Uchiyama, S, Nishina, K, Ohkubo, T, Kato, H, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-30 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly
To be Published
|
|
6ILI
 
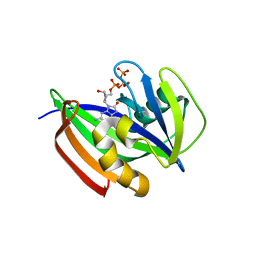 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 8-oxo-dGTP at pH 6.5 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2018-10-18 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
1X10
 
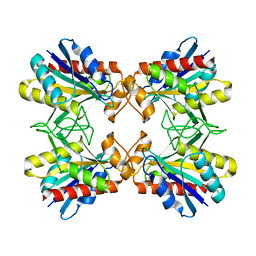 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192A) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1Z8X
 
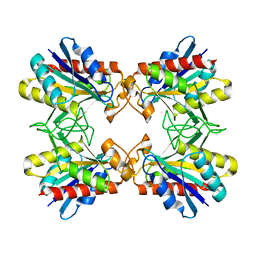 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192V) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1Z8W
 
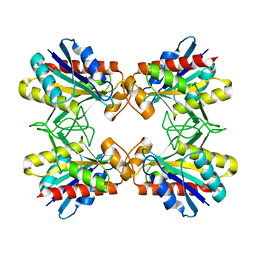 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192I) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
2DZV
 
 | |
2DZT
 
 | |
2DZU
 
 | |
7WW6
 
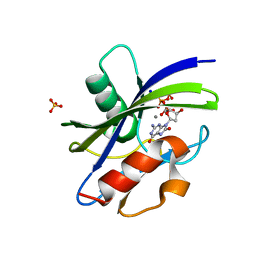 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 20 min in 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW7
 
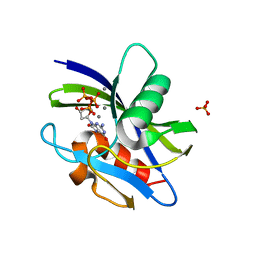 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 1 hr in 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW9
 
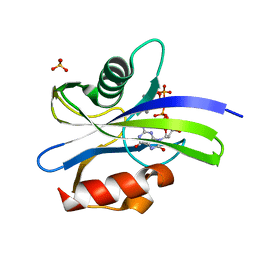 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 1.5 hr in 20 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW8
 
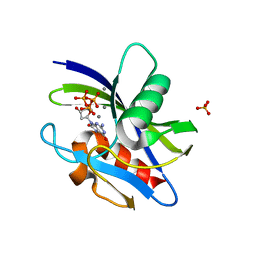 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 5 hr in 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WWA
 
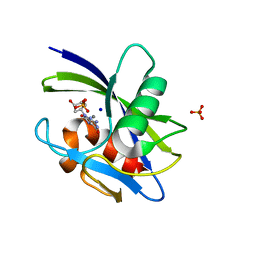 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 2.5 hr in 20 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2DZS
 
 | |
