8Z50
 
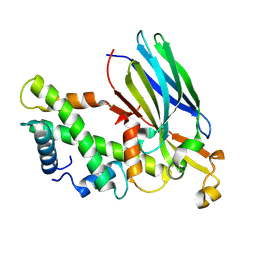 | | Crystal structure of the ASF1-H3T-H4 complex | | 分子名称: | Histone H3.1t, Histone H4, Histone chaperone ASF1A | | 著者 | Xu, L. | | 登録日 | 2024-04-18 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into instability of the nucleosome driven by histone variant H3T.
Biochem.Biophys.Res.Commun., 727, 2024
|
|
2ODM
 
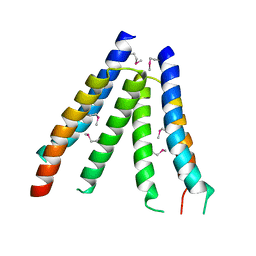 | | Crystal structure of S. aureus YlaN, an essential leucine rich protein involved in the control of cell shape | | 分子名称: | UPF0358 protein MW0995 | | 著者 | Xu, L, Sedelnikova, S.E, Baker, P.J, Errington, J, Hunt, A, Rice, D.W. | | 登録日 | 2006-12-23 | | 公開日 | 2007-06-05 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Crystal structure of S. aureus YlaN, an essential leucine rich protein involved in the control of cell shape.
Proteins, 68, 2007
|
|
8WO7
 
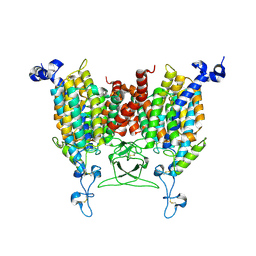 | | Apo state of Arabidopsis AZG1 T440Y | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | 著者 | Xu, L, Guo, J. | | 登録日 | 2023-10-06 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8WMQ
 
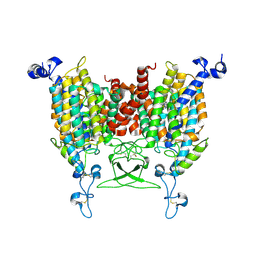 | | trans-Zeatin bound state of Arabidopsis AZG1 at pH5.5 | | 分子名称: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | 著者 | Xu, L, Guo, J. | | 登録日 | 2023-10-04 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRL
 
 | | Apo state of Arabidopsis AZG1 at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | 著者 | Xu, L, Guo, J. | | 登録日 | 2023-03-19 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRN
 
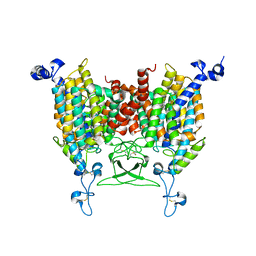 | | 6-BAP bound state of Arabidopsis AZG1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-BENZYL-9H-PURIN-6-AMINE | | 著者 | Xu, L, Guo, J. | | 登録日 | 2023-03-19 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRP
 
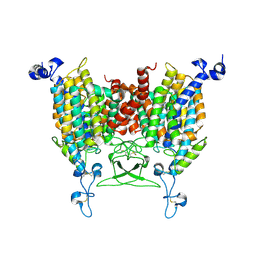 | | kinetin bound state of Arabidopsis AZG1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE | | 著者 | Xu, L, Guo, J. | | 登録日 | 2023-03-19 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRM
 
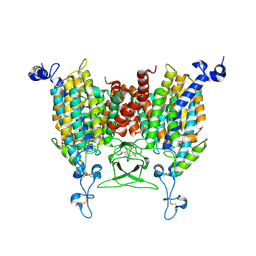 | |
8IRO
 
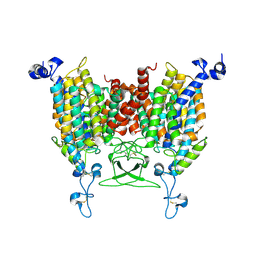 | | trans-Zeatin bound state of Arabidopsis AZG1 at pH7.4 | | 分子名称: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | 著者 | Xu, L, Guo, J. | | 登録日 | 2023-03-19 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
7WIF
 
 | | The THF-II riboswitch bound to H4B | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, RNA (50-MER) | | 著者 | Xu, L, Fang, X, Xiao, Y. | | 登録日 | 2022-01-03 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7WIE
 
 | | The THF-II riboswitch bound to 7DG | | 分子名称: | 7-DEAZAGUANINE, RNA (50-MER) | | 著者 | Xu, L, Fang, X, Xiao, Y. | | 登録日 | 2022-01-03 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7WIA
 
 | |
7WII
 
 | | The THF-II riboswitch bound to NPR | | 分子名称: | 2-AMINO-7,8-DIHYDRO-6-(1,2,3-TRIHYDROXYPROPYL)-4(1H)-PTERIDINONE, RNA (50-MER) | | 著者 | Xu, L, Fang, X, Xiao, Y. | | 登録日 | 2022-01-03 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7WI9
 
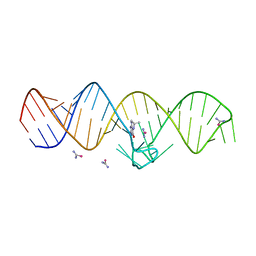 | | The THF-II riboswitch bound to THF and soaking with SeUrea | | 分子名称: | (6S)-5,6,7,8-TETRAHYDROFOLATE, RNA (50-MER), selenourea | | 著者 | Xu, L, Fang, X, Xiao, Y. | | 登録日 | 2022-01-03 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7WIB
 
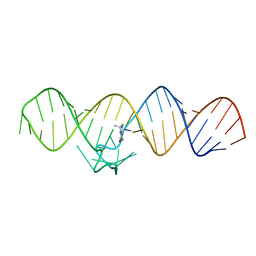 | | The THF-II riboswitch bound to THF | | 分子名称: | (6S)-5,6,7,8-TETRAHYDROFOLATE, RNA (50-MER) | | 著者 | Xu, L, Fang, X, Xiao, Y. | | 登録日 | 2022-01-03 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7DB7
 
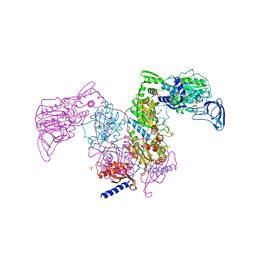 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase in complex with compound GDI05-001 | | 分子名称: | 1-[3-[2-(1H-indol-3-yl)ethylsulfamoyl]phenyl]-3-(1,3-thiazol-2-yl)urea, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, ... | | 著者 | Xu, M, Zhang, X, Xu, L, Chen, S. | | 登録日 | 2020-10-19 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7DAW
 
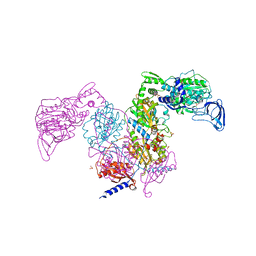 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase | | 分子名称: | Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, SULFATE ION | | 著者 | Xu, M, Zhang, X, Xu, L, Chen, S. | | 登録日 | 2020-10-18 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7DB8
 
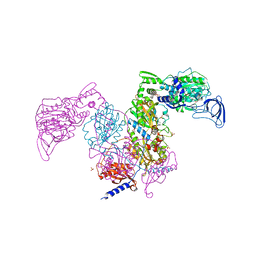 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase in complex with compound PF-3845 | | 分子名称: | N-pyridin-3-yl-4-[[3-[5-(trifluoromethyl)pyridin-2-yl]oxyphenyl]methyl]piperidine-1-carboxamide, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, ... | | 著者 | Xu, M, Zhang, X, Xu, L, Chen, S. | | 登録日 | 2020-10-19 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
8IOY
 
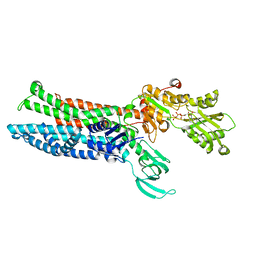 | | Structure of ATP7B C983S/C985S/D1027A mutant with AMP-PNP | | 分子名称: | Copper-transporting ATPase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Yang, G, Xu, L, Guo, J, Wu, Z. | | 登録日 | 2023-03-13 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | 分子名称: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | 登録日 | 2020-12-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
6NTY
 
 | | 2.1 A resolution structure of the Musashi-2 (Msi2) RNA recognition motif 1 (RRM1) domain | | 分子名称: | PHOSPHATE ION, RNA-binding protein Musashi homolog 2 | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Lan, L, Xiaoqing, W, Cooper, A, Gao, F.P, Xu, L. | | 登録日 | 2019-01-30 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 88, 2020
|
|
