6QYD
 
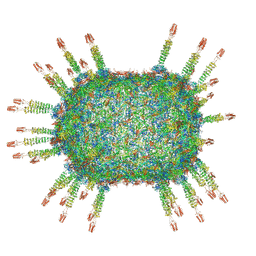 | | Cryo-EM structure of the head in mature bacteriophage phi29 | | Descriptor: | Capsid fiber protein, Major capsid protein | | Authors: | Xu, J.W, Wang, D.H, Gui, M, Xiang, Y. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural assembly of the tailed bacteriophage φ29.
Nat Commun, 10, 2019
|
|
6QVK
 
 | |
6QZ0
 
 | | The cryo-EM structure of the head of the genome empited bacteriophage phi29 | | Descriptor: | Capsid fiber protein, Major capsid protein | | Authors: | Xu, J, Wang, D, Gui, M, Xiang, Y. | | Deposit date: | 2019-03-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural assembly of the tailed bacteriophage φ29.
Nat Commun, 10, 2019
|
|
7DRX
 
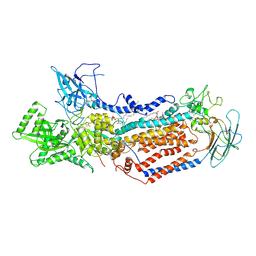 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with beryllium fluoride (E2P state) | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-30 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DSH
 
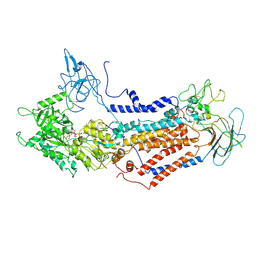 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with AMPPCP (E1-ATP state) | | Descriptor: | Alkylphosphocholine resistance protein LEM3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DSI
 
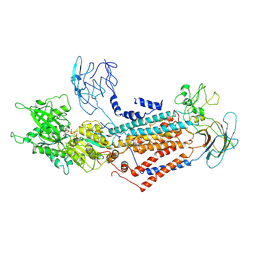 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in yeast lipids with AMPPCP ( resting state ) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
5GW8
 
 | | Crystal structure of a putative DAG-like lipase (MgMDL2) from Malassezia globosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Xu, J, Xu, H, Liu, J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Malassezia globosa MgMDL2 lipase: Crystal structure and rational modification of substrate specificity.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
4ZRD
 
 | | Crystal structure of SMG1 F278N mutant | | Descriptor: | GLYCEROL, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of product-bound SMG1 lipase: active site gating implications.
Febs J., 282, 2015
|
|
4ZRE
 
 | | Crystal structure of SMG1 F278D mutant | | Descriptor: | CHLORIDE ION, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of product-bound SMG1 lipase: active site gating implications.
Febs J., 282, 2015
|
|
3WDJ
 
 | | Crystal structure of Pullulanase complexed with maltotetraose from Anoxybacillus sp. LM18-11 | | Descriptor: | CALCIUM ION, Type I pullulanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, J, Ren, F, Huang, C.H, Zheng, Y, Zhen, J, Ko, T.P, Chen, C.C, Chan, H.C, Guo, R.T, Ma, Y, Song, H. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Cloning, Expression, Functional and Structural Studies of Pullulanase from Anoxybacillus sp. LM18-11
To be Published
|
|
3WDH
 
 | | Crystal structure of Pullulanase from Anoxybacillus sp. LM18-11 | | Descriptor: | CALCIUM ION, Type I pullulanase | | Authors: | Xu, J, Ren, F, Huang, C.H, Zheng, Y, Zhen, J, Ko, T.P, Chen, C.C, Chan, H.C, Guo, R.T, Ma, Y, Song, H. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cloning, Expression, Functional and Structural Studies of Pullulanase from Anoxybacillus sp. LM18-11
To be Published
|
|
3WDI
 
 | | Crystal structure of Pullulanase complexed with maltotriose from Anoxybacillus sp. LM18-11 | | Descriptor: | CALCIUM ION, Type I pullulanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, J, Ren, F, Huang, C.H, Zheng, Y, Zhen, J, Ko, T.P, Chen, C.C, Chan, H.C, Guo, R.T, Ma, Y, Song, H. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, Expression, Functional and Structural Studies of Pullulanase from Anoxybacillus sp. LM18-11
To be Published
|
|
6PX9
 
 | |
1G0J
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152S | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1G0M
 
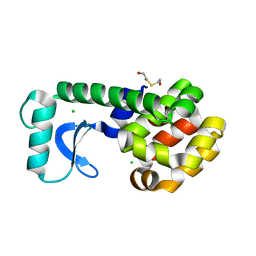 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152I | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1G0Q
 
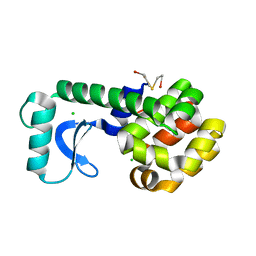 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149I | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1G0G
 
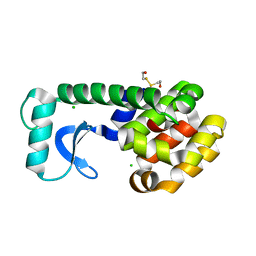 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1G0K
 
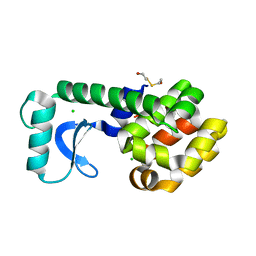 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152C | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1G06
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149S | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-05 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1G0L
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152V | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1G07
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149C | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-05 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1G0P
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149G | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
5H1B
 
 | | Human RAD51 presynaptic complex | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, MAGNESIUM ION, ... | | Authors: | Xu, J, Zhao, L, Xu, Y, Zhao, W, Sung, P, Wang, H.W. | | Deposit date: | 2016-10-08 | | Release date: | 2016-12-21 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Nat. Struct. Mol. Biol., 24, 2017
|
|
5H1C
 
 | | Human RAD51 post-synaptic complexes | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Xu, J, Zhao, L, Xu, Y, Zhao, W, Sung, P, Wang, H.W. | | Deposit date: | 2016-10-08 | | Release date: | 2016-12-21 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Nat. Struct. Mol. Biol., 24, 2017
|
|
6QZF
 
 | | The cryo-EM structure of the collar complex and tail axis in genome emptied bacteriophage phi29 | | Descriptor: | Portal protein, Pre-neck appendage protein, Proximal tail tube connector protein | | Authors: | Xu, J, Wang, D, Gui, M, Xiang, Y. | | Deposit date: | 2019-03-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural assembly of the tailed bacteriophage φ29.
Nat Commun, 10, 2019
|
|
