6N5V
 
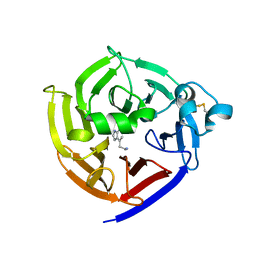 | | Crystal Structure of Strictosidine in complex with 1H-indole-4-ethanamine | | Descriptor: | 2-(1H-indol-4-yl)ethan-1-amine, Strictosidine synthase | | Authors: | Cai, Y, Shao, N, Xie, H, Futamura, Y, Panjikar, S, Liu, H, Zhu, H, Osada, H, Zou, H. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Crystal Structure of Strictosidine in complex with 1H-indole-4-ethanamine
to be published
|
|
6FV6
 
 | | Monomer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Monomer structure of Aq128 in the outward-facing state
To Be Published
|
|
6FV8
 
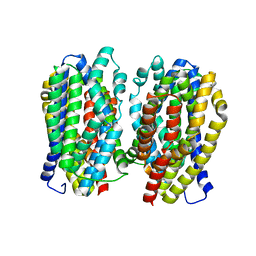 | | Dimer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dimer structure of the MATE family multidrug resistance transporter Aq128
in the outward-facing state
To Be Published
|
|
6FV7
 
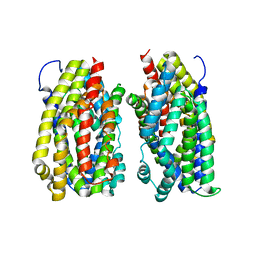 | | Dimer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Dimer structure of Aq128 in the outward-facing state
To Be Published
|
|
2XFE
 
 | | vCBM60 in complex with galactobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
2XHJ
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules. SeMet form of vCBM60. | | Descriptor: | CALCIUM ION, CALCIUM-DEPENDENT CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
7NB6
 
 | |
1YGT
 
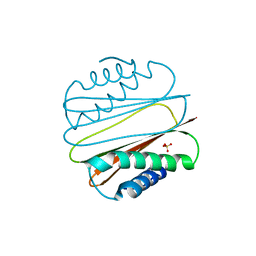 | |
2XHH
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
2XFD
 
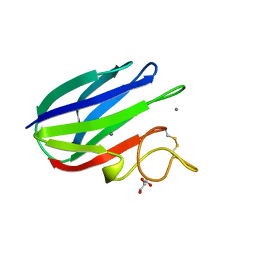 | | vCBM60 in complex with cellobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, GLYCEROL, ... | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
1US3
 
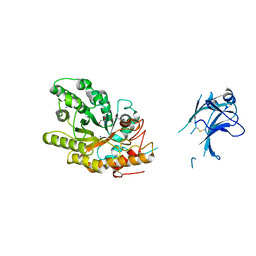 | | Native xylanase10C from Cellvibrio japonicus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-1,4-XYLANASE PRECURSOR, GLYCEROL, ... | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
1US2
 
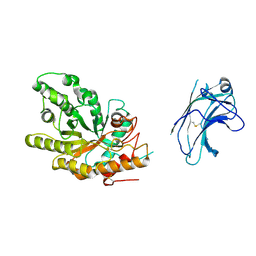 | | Xylanase10C (mutant E385A) from Cellvibrio japonicus in complex with xylopentaose | | Descriptor: | ENDO-BETA-1,4-XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
1GNY
 
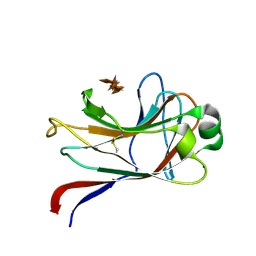 | | xylan-binding module CBM15 | | Descriptor: | SODIUM ION, XYLANASE 10C, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Szabo, S, Jamal, S, Xie, H, Charnock, S.J, Bolam, D.N, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of a Family 15 Carbohydrate-Binding Module in Complex with Xylopentaose: Evidence that Xylan Binds in an Approximate Three-Fold Helical Conformation
J.Biol.Chem., 276, 2001
|
|
5DJQ
 
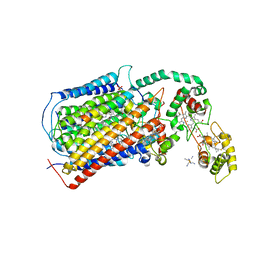 | | The structure of CBB3 cytochrome oxidase. | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoN1, ... | | Authors: | Buschmann, S, Warkentin, E, Xie, H, Kohlstaedt, M, Langer, J.D, Ermler, U, Michel, H. | | Deposit date: | 2015-09-02 | | Release date: | 2016-01-13 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of cbb3 cytochrome oxidase provides insights into proton pumping.
Science, 329, 2010
|
|
7TPR
 
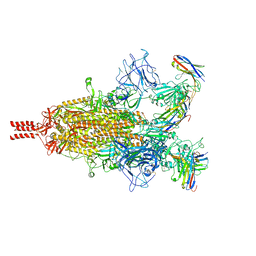 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | Descriptor: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | Authors: | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
4PUS
 
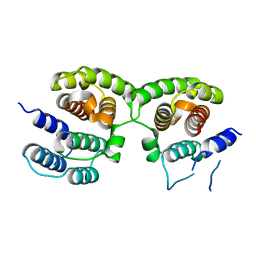 | | Crystal Structure of Influenza A Virus Matrix Protein M1 | | Descriptor: | Matrix protein 1 | | Authors: | Safo, M.K, Musayev, F.N, Mosier, P.D, Xie, H, Desai, U.R. | | Deposit date: | 2014-03-13 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of influenza a virus matrix protein m1: variations on a theme.
Plos One, 9, 2014
|
|
5V7B
 
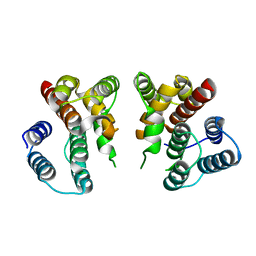 | | Crystal structure of Influenza A virus matrix protein M1 (NLS-88E) | | Descriptor: | Matrix protein 1 | | Authors: | Musayev, F.N, Safo, M.K, Desai, U.R, Xie, H, Mosier, P.D, Chiang, M.-J. | | Deposit date: | 2017-03-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Maintaining pH-dependent conformational flexibility of M1 is critical for efficient influenza A virus replication.
Emerg Microbes Infect, 6, 2017
|
|
8HI9
 
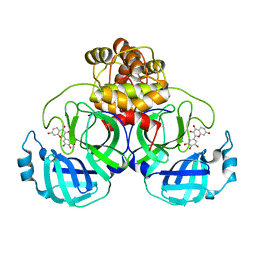 | | SARS-CoV-2 3CL protease (3CLpro) in complex with Robinetin | | Descriptor: | 3,7-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-11-19 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of Polyphenolic Natural Products as SARS-CoV-2 M pro Inhibitors for COVID-19.
Pharmaceuticals, 16, 2023
|
|
8GJM
 
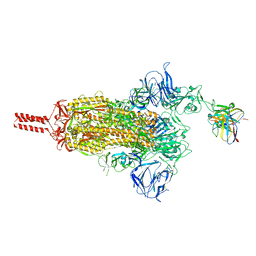 | | 17b10 fab in complex with full-length SARS-CoV-2 Spike G614 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8GJN
 
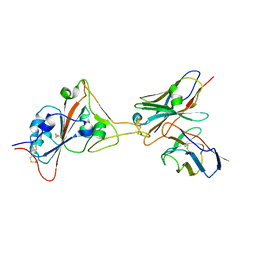 | | 17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 17B10 Fab, Light chain of 17B10 Fab, ... | | Authors: | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
6M2N
 
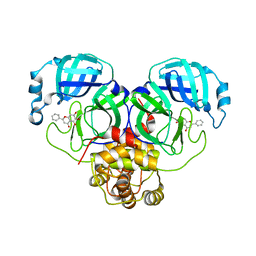 | | SARS-CoV-2 3CL protease (3CL pro) in complex with a novel inhibitor | | Descriptor: | 3C-like proteinase, 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | | Authors: | Su, H.X, Zhao, W.F, Li, M.J, Xie, H, Xu, Y.C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients.
Acta Pharmacol.Sin., 41, 2020
|
|
6M2Q
 
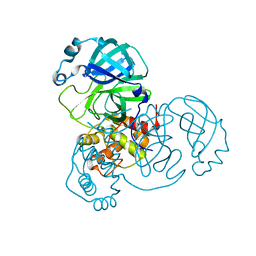 | | SARS-CoV-2 3CL protease (3CL pro) apo structure (space group C21) | | Descriptor: | 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Li, M.J, Xie, H, Xu, Y.C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients.
Acta Pharmacol.Sin., 41, 2020
|
|
7DPV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-dihydromyricetin | | Descriptor: | (2S,3S)-3,5-dihydroxy-7-methoxy-2-(3,4,5-trihydroxyphenyl)chroman-4-one, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPU
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-myricetin | | Descriptor: | 3C-like proteinase, 7-methoxy-3,5-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, GLYCEROL | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
