2Y8Q
 
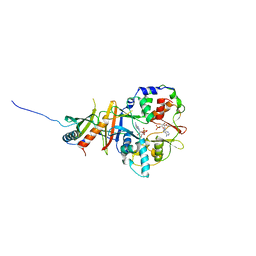 | | Structure of the regulatory fragment of mammalian AMPK in complex with one ADP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-09 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
1HKA
 
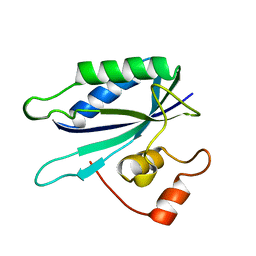 | | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Authors: | Xiao, B, Shi, G, Chen, X, Yan, H, Ji, X. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, a potential target for the development of novel antimicrobial agents.
Structure Fold.Des., 7, 1999
|
|
2Y8L
 
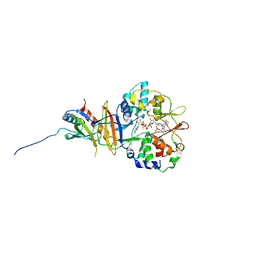 | | Structure of the regulatory fragment of mammalian aMPK in complex with two ADP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
2YA3
 
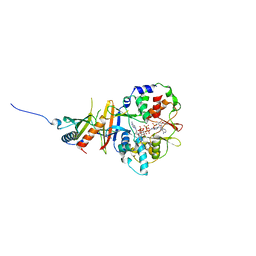 | | STRUCTURE OF THE REGULATORY FRAGMENT OF MAMMALIAN AMPK IN COMPLEX WITH COUMARIN ADP | | Descriptor: | 3'-(7-diethylaminocoumarin-3-carbonylamino)-3'-deoxy-ADP, 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
2BQZ
 
 | | Crystal structure of a ternary complex of the human histone methyltransferase Pr-SET7 (also known as SET8) | | Descriptor: | HISTONE H4, S-ADENOSYL-L-HOMOCYSTEINE, SET8 PROTEIN | | Authors: | Xiao, B, Jing, C, Kelly, G, Walker, P.A, Muskett, F.W, Frenkiel, T.A, Martin, S.R, Sarma, K, Reinberg, D, Gamblin, S.J, Wilson, J.R. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity and Mechanism of the Histone Methyltransferase Pr-Set7
Genes Dev., 19, 2005
|
|
2V92
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
2V9J
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with Mg.ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-23 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
4CFH
 
 | | Structure of an active form of mammalian AMPK | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-18 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
1QRY
 
 | | Homeobox protein VND (ventral nervous system defective protein) | | Descriptor: | PROTEIN (HOMEOBOX VENTRAL NERVOUS SYSTEM DEFECTIVE PROTEIN) | | Authors: | Xiang, B. | | Deposit date: | 1999-06-16 | | Release date: | 1999-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Distortion of the three-dimensional structure of the vnd/NK-2 homeodomain bound to DNA induced by an embryonically lethal A35T point mutation.
Biochemistry, 42, 2003
|
|
3QL9
 
 | |
3QLA
 
 | | Hexagonal complex structure of ATRX ADD bound to H3K9me3 peptide | | Descriptor: | POTASSIUM ION, Transcriptional regulator ATRX, ZINC ION, ... | | Authors: | Xiang, B, Li, H. | | Deposit date: | 2011-02-02 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
1EQM
 
 | | CRYSTAL STRUCTURE OF BINARY COMPLEX OF 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE WITH ADENOSINE-5'-DIPHOSPHATE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xiao, B, Blaszczyk, J, Ji, X. | | Deposit date: | 2000-04-05 | | Release date: | 2001-04-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual conformational changes in 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as revealed by X-ray crystallography and NMR.
J.Biol.Chem., 276, 2001
|
|
2L92
 
 | |
3BR8
 
 | |
2KYT
 
 | | Solution structure of the H-REV107 N-terminal domain | | Descriptor: | Group XVI phospholipase A2 | | Authors: | Ren, X, Xia, B. | | Deposit date: | 2010-06-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal catalytic domain of human H-REV107--a novel circular permutated NlpC/P60 domain
Febs Lett., 584, 2010
|
|
2L17
 
 | | An arsenate reductase in the reduced state | | Descriptor: | Arsenate reductase | | Authors: | Yu, C, Xia, B, Jin, C. | | Deposit date: | 2010-07-26 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | (1)H, (13)C and (15)N resonance assignments of the arsenate reductase from Synechocystis sp. strain PCC 6803
Biomol.Nmr Assign., 5, 2011
|
|
2LKT
 
 | |
2L18
 
 | |
2L19
 
 | | An arsenate reductase in the intermediate state | | Descriptor: | Arsenate reductase | | Authors: | Yu, C, Xia, B, Jin, C. | | Deposit date: | 2010-07-26 | | Release date: | 2011-04-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | (1)H, (13)C and (15)N resonance assignments of the arsenate reductase from Synechocystis sp. strain PCC 6803
Biomol.Nmr Assign., 5, 2011
|
|
2B5X
 
 | |
2B5Y
 
 | |
2B8G
 
 | |
6E4H
 
 | | Solution NMR Structure of the Colied-coil PALB2 Homodimer | | Descriptor: | Partner and localizer of BRCA2 | | Authors: | Song, F, Li, M, Liu, G, Swapna, G.V.T, Xia, B, Bunting, S.F, Montelione, G.T. | | Deposit date: | 2018-07-17 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antiparallel Coiled-Coil Interactions Mediate the Homodimerization of the DNA Damage-Repair Protein PALB2.
Biochemistry, 57, 2018
|
|
2AI6
 
 | |
1Z6H
 
 | | Solution Structure of Bacillus subtilis BLAP biotinylated-form | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Biotin/Lipoyl Attachment Protein | | Authors: | Cui, G, Xia, B. | | Deposit date: | 2005-03-22 | | Release date: | 2006-03-22 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Identification and solution structures of a single domain biotin/lipoyl attachment protein from Bacillus subtilis
J.Biol.Chem., 281, 2006
|
|
