2LLG
 
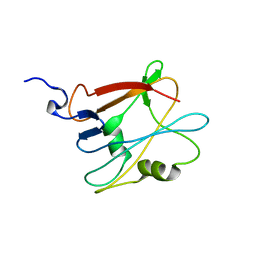 | | NMR structure of the protein NP_814968.1 from Enterococcus faecalis | | Descriptor: | Uncharacterized protein | | Authors: | Susac, L, Serrano, P, Geralt, M, Mohanty, B, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2011-11-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NP_814968.1 from Enterococcus faecalis
To be Published
|
|
2MHN
 
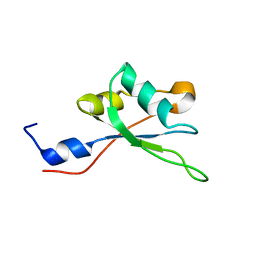 | |
2M2B
 
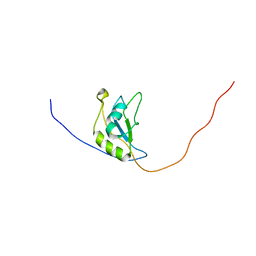 | | NMR structure of the RRM2 domain of the protein RBM10 from Homo sapiens | | Descriptor: | RNA-binding protein 10 | | Authors: | Serrano, P, Geralt, M, Dutta, S.K, Wuthrich, K, Wrobel, R.L, Makino, S, Misenhiemer, T.M, Markley, J.L, Fox, B.G, Joint Center for Structural Genomics (JCSG), Partnership for T-Cell Biology (TCELL), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the RRM2 domain of the protein RBM10 from Homo sapiens
To be Published
|
|
2N2F
 
 | | Solution NMR structure of Dynorphin 1-13 bound to Kappa Opioid Receptor | | Descriptor: | Dynorphin A(1-13) | | Authors: | O'Connor, C, White, K, Doncescu, N, Didenko, T, Roth, B.L, Czaplicki, G, Stevens, R.C, Wuthrich, K, Milon, A. | | Deposit date: | 2015-05-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2KA7
 
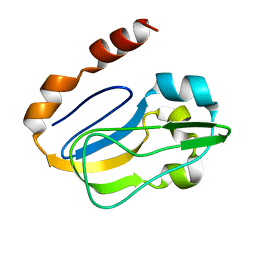 | | NMR solution structure of TM0212 at 40 C | | Descriptor: | Glycine cleavage system H protein | | Authors: | Pedrini, B, Herrmann, T, Mohanty, B, Geralt, M, Wilson, I, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The J-UNIO protocol for automated protein structure determination by NMR in solution.
J.Biomol.Nmr, 53, 2012
|
|
2MHL
 
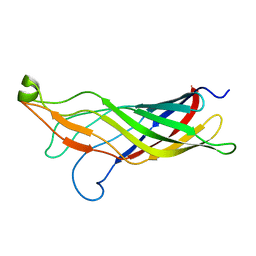 | |
2MDZ
 
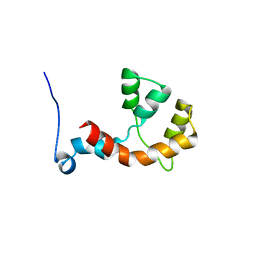 | | NMR structure of the Paracoccus denitrificans Z-subunit determined in the presence of ADP | | Descriptor: | Uncharacterized protein | | Authors: | Serrano, P, Geralt, M, Wuthrich, K, Morales-Rios, E, Zarco-Zavala, M, Garcia-Trejo, J.J, Dutta, S.K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the putative ATPase regulatory protein YP_916642.1 from Paracoccus denitrificans
To be Published
|
|
2N6D
 
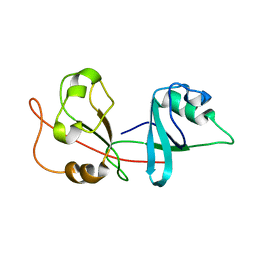 | |
2MXT
 
 | |
2NSW
 
 | |
2NBB
 
 | |
2FNB
 
 | | NMR STRUCTURE OF THE FIBRONECTIN ED-B DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (FIBRONECTIN) | | Authors: | Fattorusso, R, Pellecchia, M, Viti, F, Neri, P, Neri, D, Wuthrich, K. | | Deposit date: | 1998-12-16 | | Release date: | 1998-12-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human oncofoetal fibronectin ED-B domain, a specific marker for angiogenesis.
Structure Fold.Des., 7, 1999
|
|
2NSV
 
 | |
2N8G
 
 | | NMR Structure of the homeodomain transcription factor Gbx1[E23R,R58E] from Homo sapiens | | Descriptor: | Homeobox protein GBX-1 | | Authors: | Proudfoot, A.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2015-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the homeodomain transcription factor Gbx1[E23R,R58E] from Homo sapiens
To be Published
|
|
2N6E
 
 | |
2JMR
 
 | | NMR structure of the E. coli type 1 pilus subunit FimF | | Descriptor: | fimF | | Authors: | Gossert, A.D, Bettendorff, P, Puorger, C, Vetsch, M, Herrmann, T, Fiorito, F, Hiller, S, Glockshuber, R, Wuthrich, K. | | Deposit date: | 2006-11-29 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Escherichia coli type 1 pilus subunit FimF and its interactions with other pilus subunits.
J.Mol.Biol., 375, 2008
|
|
2IDY
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Horst, R, Herrmann, T, Joseph, J, Saikatendu, K, Subramanian, V, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2GDT
 
 | | NMR Structure of the nonstructural protein 1 (nsp1) from the SARS coronavirus | | Descriptor: | Leader protein; p65 homolog; NSP1 (EC 3.4.22.-) | | Authors: | Almeida, M.S, Herrmann, T, Geralt, M, Johnson, M.A, Saikatendu, K, Joseph, J, Subramanian, R.C, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-03-17 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2G9O
 
 | | Solution structure of the apo form of the third metal-binding domain of ATP7A protein (Menkes Disease protein) | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
2GRI
 
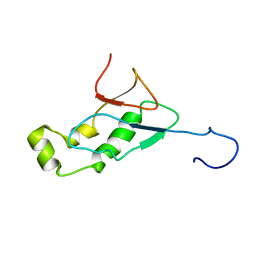 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Herrmann, T, Saikatendu, K.S, Joseph, J, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2GA7
 
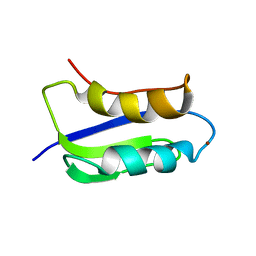 | | Solution structure of the copper(I) form of the third metal-binding domain of ATP7A protein (menkes disease protein) | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
2I3B
 
 | |
2KC6
 
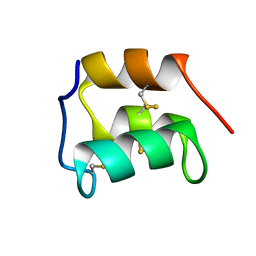 | | NMR solution structure of the pheromone En-1 of Euplotes nobilii at -1.5 C | | Descriptor: | Mating pheromone En-1 | | Authors: | Pedrini, B, Alimenti, C, Vallesi, A, Luporini, P, Wuthrich, K. | | Deposit date: | 2008-12-17 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Molecular cold-adaptation: Comparative analysis of two homologous families of psychrophilic and mesophilic signal proteins of the protozoan ciliate, Euplotes.
Iubmb Life, 61, 2009
|
|
