6KXT
 
 | | BON1-C2B | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Protein BONZAI 1, SULFATE ION | | Authors: | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
6LUO
 
 | | Structure of nurse shark beta-2-microglobulin | | Descriptor: | Beta-2-microglobulin | | Authors: | Xia, C, Wu, Y. | | Deposit date: | 2020-01-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between beta 2 -Microglobulin and H Chain Conserved in Evolution.
J Immunol., 2021
|
|
6MHN
 
 | | Photoactive Yellow Protein with covalently bound 3-chloro-4-hydroxycinnamic acid chromophore | | Descriptor: | (2E)-3-(3-chloro-4-hydroxyphenyl)prop-2-enoic acid, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
6MKT
 
 | | Photoactive Yellow Protein with 3-chlorotyrosine substituted at position 42 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
6MMD
 
 | | Photoactive Yellow Protein with 3,5-dichlorotyrosine substituted at position 42 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-30 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.228 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
7VIV
 
 | |
5V57
 
 | | 3.0A SYN structure of the multi-domain human smoothened receptor in complex with TC114 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FLAVIN MONONUCLEOTIDE, N-methyl-N-[1-[4-(2-methylpyrazol-3-yl)phthalazin-1-yl]piperidin-4-yl]-4-nitro-2-(trifluoromethyl)benzamide, ... | | Authors: | Zhang, X, Zhao, F, Wu, Y, Yang, J, Han, G.W, Zhao, S, Ishchenko, A, Ye, L, Lin, X, Ding, K, Dharmarajan, V, Griffin, P.R, Gati, C, Nelson, G, Hunter, M.S, Hanson, M.A, Cherezov, V, Stevens, R.C, Tan, W, Tao, H, Xu, F. | | Deposit date: | 2017-03-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand.
Nat Commun, 8, 2017
|
|
5V56
 
 | | 2.9A XFEL structure of the multi-domain human smoothened receptor (with E194M mutation) in complex with TC114 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN MONONUCLEOTIDE, N-methyl-N-[1-[4-(2-methylpyrazol-3-yl)phthalazin-1-yl]piperidin-4-yl]-4-nitro-2-(trifluoromethyl)benzamide, ... | | Authors: | Zhang, X, Zhao, F, Wu, Y, Yang, J, Han, G.W, Zhao, S, Ishchenko, A, Ye, L, Lin, X, Ding, K, Dharmarajan, V, Griffin, P.R, Gati, C, Nelson, G, Hunter, M.S, Hanson, M.A, Cherezov, V, Stevens, R.C, Tan, W, Tao, H, Xu, F. | | Deposit date: | 2017-03-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand.
Nat Commun, 8, 2017
|
|
8YZ5
 
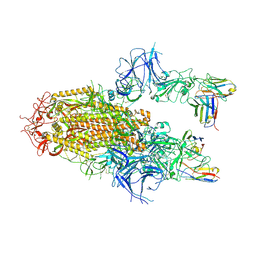 | | SARS-CoV-2 Delta Spike in complex with Fab of JE-5C | | Descriptor: | Fab heavy chain of JE-5C, Fab light chain of JE-5C, Spike glycoprotein | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2024-04-06 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine
Plos Pathog., 2024
|
|
8YZ6
 
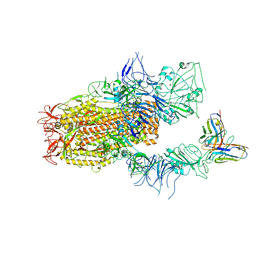 | | SARS-CoV-2 Spike (BA.1) in complex with Fab of JH-8B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain of JH-8B, ... | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2024-04-06 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine
Plos Pathog., 2024
|
|
5TGY
 
 | | NMR structure of holo-PS1 | | Descriptor: | PS1, [5,10,15,20-tetrakis(trifluoromethyl)porphyrinato(2-)-kappa~4~N~21~,N~22~,N~23~,N~24~]zinc | | Authors: | Polizzi, N.F, Wu, Y. | | Deposit date: | 2016-09-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of a hyperstable non-natural protein-ligand complex with sub- angstrom accuracy.
Nat Chem, 9, 2017
|
|
5TGW
 
 | | NMR structure of apo-PS1 | | Descriptor: | PS1 | | Authors: | Polizzi, N.F, Wu, Y. | | Deposit date: | 2016-09-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of a hyperstable non-natural protein-ligand complex with sub- angstrom accuracy.
Nat Chem, 9, 2017
|
|
7FGC
 
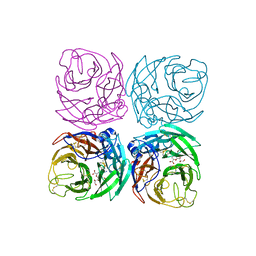 | |
7FGD
 
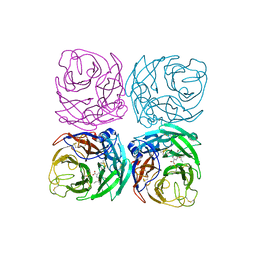 | | A naturally-occurring neuraminidase-inhibitors-resistant NA from asiatic toad influenza B-like virus | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, toad NA (OSE) | | Authors: | Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2021-07-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and inhibitor sensitivity analysis of influenza B-like viral neuraminidases derived from Asiatic toad and spiny eel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7FGB
 
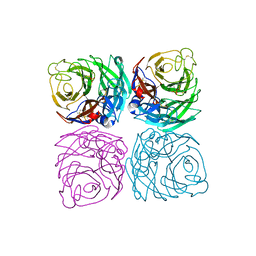 | | A naturally-occurring neuraminidase-inhibitors-resistant NA from asiatic toad influenza B-like virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | Authors: | Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2021-07-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and inhibitor sensitivity analysis of influenza B-like viral neuraminidases derived from Asiatic toad and spiny eel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4LMM
 
 | | Crystal structure of NHERF1 PDZ1 domain complexed with the CXCR2 C-terminal tail in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-07-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
7FGE
 
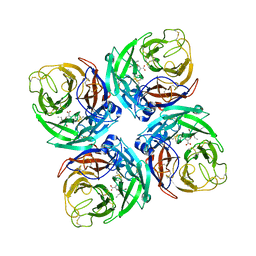 | |
4N6X
 
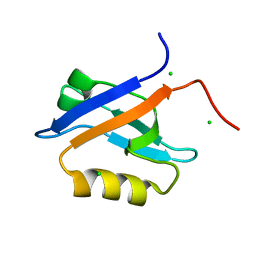 | | Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1/Chemokine Receptor CXCR2 fusion protein | | Authors: | Lu, G, Wu, Y, Jiang, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.051 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
4MPA
 
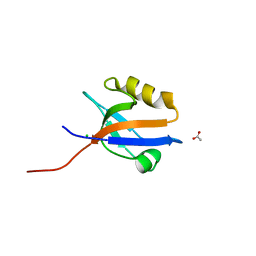 | | Crystal structure of NHERF1-CXCR2 signaling complex in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1, ... | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
2JS4
 
 | | Solution NMR Structure of Bordetella bronchiseptica protein BB2007. Northeast Structural Genomics Consortium target BoR54 | | Descriptor: | UPF0434 protein BB2007 | | Authors: | Eletsky, A, Sukumaran, D, Wu, Y, Singarapu, K, Parish, D, Xu, D, Wang, D, Nwosu, C, Cunningham, K, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Bordetella bronchiseptica protein BB2007.
To be Published
|
|
5ZKT
 
 | |
