1M2M
 
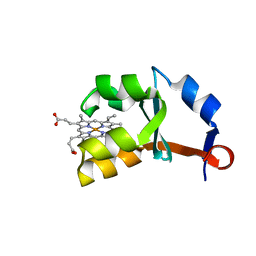 | | Crystal structure of E44A/E48A/E56A/D60A mutant of cytochrome b5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Wu, J, Wang, Y.-H, Gan, J.-H, Wang, W.-H, Sun, B.-Y, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2002-06-24 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Cytochrome b5 Mutated at the Charged Surface-Residues and Their Interactions with Cytochrome c
Chin.J.Chem., 20, 2002
|
|
3H0R
 
 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASPARAGINE, ... | | Authors: | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | Deposit date: | 2009-04-10 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
1EHB
 
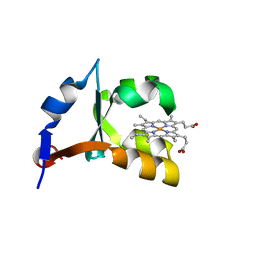 | | CRYSTAL STRUCTURE OF RECOMBINANT TRYPSIN-SOLUBILIZED FRAGMENT OF CYTOCHROME B5 | | Descriptor: | PROTEIN (CYTOCHROME B5), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, J, Gan, J.-H, Xia, Z.-X, Wang, Y.-H, Wang, W.-H, Xue, L.-L, Xie, Y, Huang, Z.-X. | | Deposit date: | 2000-02-20 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of recombinant trypsin-solubilized fragment of cytochrome b(5) and the structural comparison with Val61His mutant.
Proteins, 40, 2000
|
|
3H0M
 
 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, ... | | Authors: | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | Deposit date: | 2009-04-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
3H0L
 
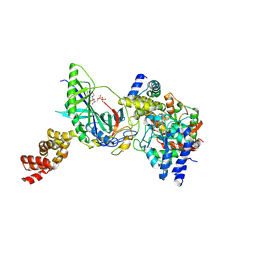 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ASPARAGINE, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, ... | | Authors: | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | Deposit date: | 2009-04-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
7VCF
 
 | | Cryo-EM structure of Chlamydomonas TOC-TIC supercomplex | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, J, Yan, Z, Jin, Z, Zhang, Y. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of a TOC-TIC supercomplex spanning two chloroplast envelope membranes.
Cell, 185, 2022
|
|
3TCA
 
 | |
4Q9V
 
 | | Crystal structure of TIPE3 | | Descriptor: | CHLORIDE ION, SULFATE ION, Tumor necrosis factor alpha-induced protein 8-like protein 3 | | Authors: | Wu, J, Zhang, X, Chen, Y.H, Shi, Y. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer.
Cancer Cell, 26, 2014
|
|
1ES1
 
 | | CRYSTAL STRUCTURE OF VAL61HIS MUTANT OF TRYPSIN-SOLUBILIZED FRAGMENT OF CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, J, Gan, J.-H, Xia, Z.-X, Wang, Y.-H, Wang, W.-H, Xue, L.-L, Xie, Y, Huang, Z.-X. | | Deposit date: | 2000-04-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of recombinant trypsin-solubilized fragment of cytochrome b(5) and the structural comparison with Val61His mutant.
Proteins, 40, 2000
|
|
2P6E
 
 | |
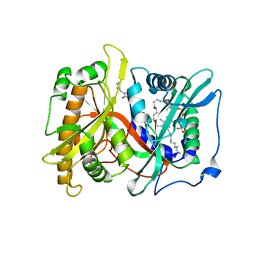
2P6G
 
 | |
2P6F
 
 | |
1NE4
 
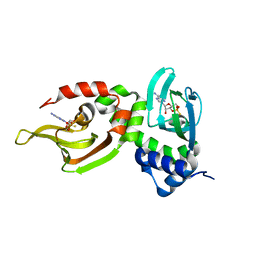 | | Crystal Structure of Rp-cAMP Binding R1a Subunit of cAMP-dependent Protein Kinase | | Descriptor: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Jones, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2002-12-10 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of RIalpha Subunit of Cyclic Adenosine 5'-Monophosphate (cAMP)-Dependent Protein Kinase Complexed with (R(p))-Adenosine 3',5'-Cyclic Monophosphothioate and (S(p))-Adenosine 3',5'-Cyclic Monophosphothioate, the Phosphothioate Analogues of cAMP.
Biochemistry, 43, 2004
|
|
5IC0
 
 | | Structural analysis of a talin triple domain module | | Descriptor: | 1,2-ETHANEDIOL, Talin-1 | | Authors: | Wu, J, Chang, Y.-C.E, Zhang, H, Brennan, M.L. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration.
Structure, 24, 2016
|
|
5IC1
 
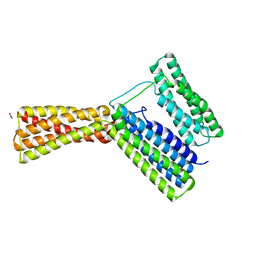 | | Structural analysis of a talin triple domain module, E1794Y, E1797Y, Q1801Y mutant | | Descriptor: | 1,2-ETHANEDIOL, Talin-1 | | Authors: | Wu, J, Chang, Y.-C.E, Zhang, H, Huang, Q.-Q. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration.
Structure, 24, 2016
|
|
1M2I
 
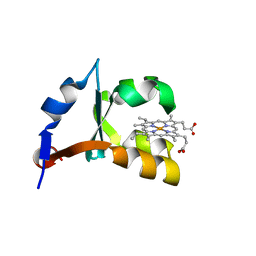 | | Crystal structure of E44A/E56A mutant of cytochrome b5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Wu, J, Wang, Y.-H, Gan, J.-H, Wang, W.-H, Sun, B.-Y, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2002-06-24 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Cytochrome b5 Mutated at the Charged Surface-Residues and Their Interactions with Cytochrome c
Chin.J.Chem., 20, 2002
|
|
1NE6
 
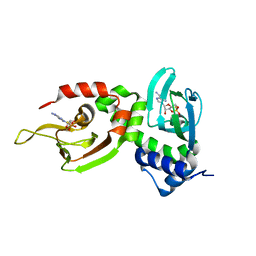 | | Crystal structure of Sp-cAMP binding R1a subunit of cAMP-dependent protein kinase | | Descriptor: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Jones, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2002-12-10 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of RIalpha Subunit of Cyclic Adenosine 5'-Monophosphate (cAMP)-Dependent Protein Kinase Complexed with (R(p))-Adenosine 3',5'-Cyclic Monophosphothioate and (S(p))-Adenosine 3',5'-Cyclic Monophosphothioate, the Phosphothioate Analogues of cAMP.
Biochemistry, 43, 2004
|
|
7CM3
 
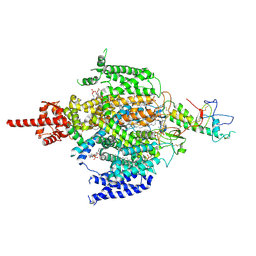 | | Cryo-EM structure of human NALCN in complex with FAM155A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J, Yan, Z, Ke, M. | | Deposit date: | 2020-07-24 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human sodium leak channel NALCN in complex with FAM155A.
Nat Commun, 11, 2020
|
|
4IBM
 
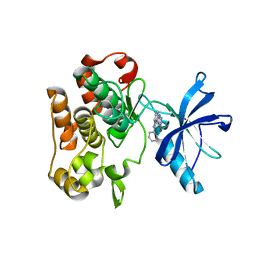 | | Crystal structure of insulin receptor kinase domain in complex with an inhibitor Irfin-1 | | Descriptor: | 5-(2-phenylpyrazolo[1,5-a]pyridin-3-yl)-3H-pyrazolo[3,4-c]pyridazin-3-one, Insulin receptor | | Authors: | Wu, J, Anastassiadis, T, Duong-Ly, K.C, Peterson, J.R. | | Deposit date: | 2012-12-08 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A highly selective dual insulin receptor (IR)/insulin-like growth factor 1 receptor (IGF-1R) inhibitor derived from an extracellular signal-regulated kinase (ERK) inhibitor.
J.Biol.Chem., 288, 2013
|
|
6AHV
 
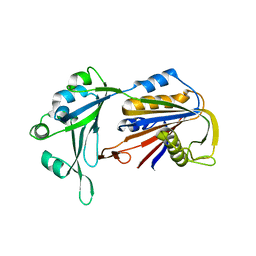 | | Crystal structure of human RPP40 | | Descriptor: | Ribonuclease P protein subunit p40 | | Authors: | Wu, J, Niu, S, Tan, M, Lan, P, Lei, M. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Cell, 175, 2018
|
|
6AHU
 
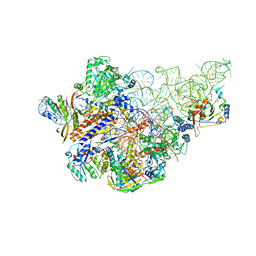 | | Cryo-EM structure of human Ribonuclease P with mature tRNA | | Descriptor: | H1 RNA, Ribonuclease P protein subunit p14, Ribonuclease P protein subunit p20, ... | | Authors: | Wu, J, Niu, S, Tan, M, Lan, P, Lei, M. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Cell, 175, 2018
|
|
6AHR
 
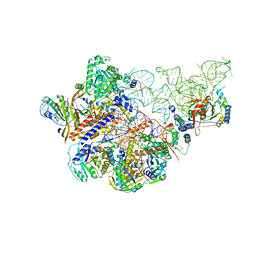 | | Cryo-EM structure of human Ribonuclease P | | Descriptor: | H1 RNA, Ribonuclease P protein subunit p14, Ribonuclease P protein subunit p20, ... | | Authors: | Wu, J, Niu, S, Tan, M, Lan, P, Lei, M. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Cell, 175, 2018
|
|
7DLU
 
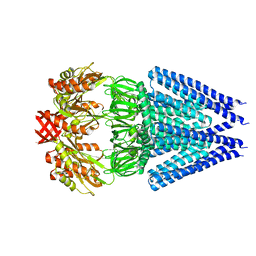 | | Mechanosensitive channel MscS K180R mutant | | Descriptor: | Mechanosensitive ion channel MscS | | Authors: | Wu, J, Ke, M. | | Deposit date: | 2020-11-30 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanosensitive channel MscS K180R mutant
To Be Published
|
|
4W8P
 
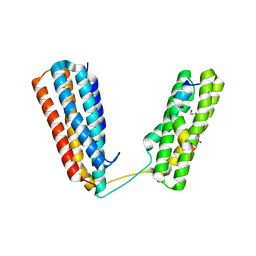 | | Crystal structure of RIAM TBS1 in complex with talin R7R8 domains | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta A4 precursor protein-binding family B member 1-interacting protein, Talin-1 | | Authors: | Chang, Y.C.E, Zhang, H, Wu, J. | | Deposit date: | 2014-08-25 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Mechanistic Insights into the Recruitment of Talin by RIAM in Integrin Signaling.
Structure, 22, 2014
|
|
2AIN
 
 | |
