2JXN
 
 | | Solution Structure of S. cerevisiae PDCD5-like Protein Ymr074cp | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Uncharacterized protein YMR074C | | Authors: | Hong, J, Zhang, J, Liu, Z, Shi, Y, Wu, J. | | Deposit date: | 2007-11-23 | | Release date: | 2008-12-02 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of S. cerevisiae PDCD5-like Protein Ymr074cp Determined by Heteronuclear NMR Spectroscopy
To be Published
|
|
2JXW
 
 | |
2H60
 
 | | Solution Structure of Human Brg1 Bromodomain | | Descriptor: | Probable global transcription activator SNF2L4 | | Authors: | Shen, W, Xu, C, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2006-05-30 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails
Biochemistry, 46, 2007
|
|
1Q2K
 
 | | Solution structure of BmBKTx1 a new potassium channel blocker from the Chinese Scorpion Buthus martensi Karsch | | Descriptor: | Neurotoxin BmK37 | | Authors: | Cai, Z, Xu, C, Xu, Y, Lu, W, Chi, C.W, Shi, Y, Wu, J. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmBKTx1, a New BK(Ca)(1) Channel Blocker from the Chinese Scorpion Buthus martensi Karsch(,).
Biochemistry, 43, 2004
|
|
3T7J
 
 | | Crystal structure of Rtt107p (residues 820-1070) | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
4OST
 
 | | Crystal structure of the S505C mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
1SJ6
 
 | | NMR Structure and Regulated Expression in APL Cell of Human SH3BGRL3 | | Descriptor: | SH3 domain-binding glutamic acid-rich-like protein 3 | | Authors: | Xu, C, Tang, Y, Xu, Y, Wu, J, Shi, Y, Zhang, Q, Zheng, P, Du, Y. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure and regulated expression in APL cell of human SH3BGRL3.
Febs Lett., 579, 2005
|
|
3DAE
 
 | |
3DIT
 
 | |
4ZOU
 
 | |
2M80
 
 | | Solution structure of yeast dithiol glutaredoxin Grx8 | | Descriptor: | Glutaredoxin-8 | | Authors: | Tang, Y, Zhang, J, Yu, J, Wu, J, Zhou, C.Z, Shi, Y. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-guided activity enhancement and catalytic mechanism of yeast grx8
Biochemistry, 53, 2014
|
|
3Q53
 
 | |
4LNQ
 
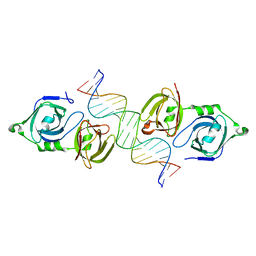 | |
5XSY
 
 | | Structure of the Nav1.4-beta1 complex from electric eel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein, Voltage-gated sodium channel beta subunit 1, ... | | Authors: | Yan, Z, Zhou, Q, Wu, J.P, Yan, N. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Nav1.4-beta 1 Complex from Electric Eel.
Cell, 170, 2017
|
|
3NPH
 
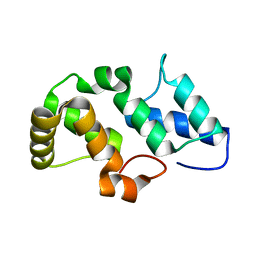 | | Crystal structure of the pfam00427 domain from Synechocystis sp. PCC 6803 | | Descriptor: | Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod 2 | | Authors: | Gao, X, Chen, L, Wu, J.-W, Zhang, Y.-Z. | | Deposit date: | 2010-06-28 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Crystal structure of the N-terminal domain of linker L(R) and the assembly of cyanobacterial phycobilisome rods
Mol.Microbiol., 82, 2011
|
|
6AH3
 
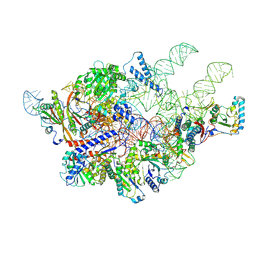 | | Cryo-EM structure of yeast Ribonuclease P with pre-tRNA substrate | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
4Q2E
 
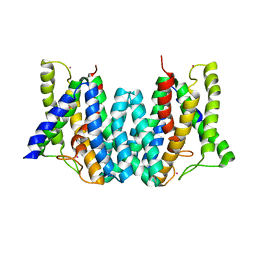 | | CRYSTAL STRUCTURE OF AN INTRAMEMBRANE CDP-DAG SYNTHETASE CENTRAL FOR PHOSPHOLIPID BIOSYNTHESIS (S200C/S258C, active mutant) | | Descriptor: | MAGNESIUM ION, MERCURY (II) ION, POTASSIUM ION, ... | | Authors: | Liu, X, Yin, Y, Wu, J, Liu, Z. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis
Nat Commun, 5, 2014
|
|
6AGB
 
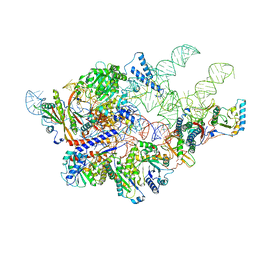 | | Cryo-EM structure of yeast Ribonuclease P | | Descriptor: | RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, Ribonuclease P protein subunit RPR2, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
1TMW
 
 | | Solution structure of Human Coactosin Like Protein D123N | | Descriptor: | Coactosin-like protein | | Authors: | Dai, H, Wu, J, Xu, Y, Tang, Y, Ding, H, Shi, Y. | | Deposit date: | 2004-06-11 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Study on Solution Structure and Its binding function to F-actin
To be Published
|
|
4Q2G
 
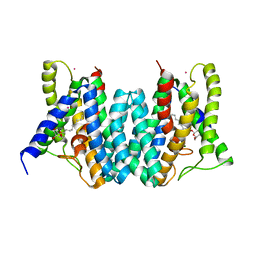 | | CRYSTAL STRUCTURE OF AN INTRAMEMBRANE CDP-DAG SYNTHETASE CENTRAL FOR PHOSPHOLIPID BIOSYNTHESIS (S200C/S223C, inactive mutant) | | Descriptor: | MAGNESIUM ION, MERCURY (II) ION, Phosphatidate cytidylyltransferase, ... | | Authors: | Liu, X, Yin, Y, Wu, J, Liu, Z. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis
Nat Commun, 5, 2014
|
|
1ERY
 
 | | PHEROMONE ER-11, NMR | | Descriptor: | PHEROMONE ER-11 | | Authors: | Luginbuhl, P, Wu, J, Zerbe, O, Ortenzi, C, Luporini, P, Wuthrich, K. | | Deposit date: | 1996-03-22 | | Release date: | 1996-08-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the pheromone Er-11 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 5, 1996
|
|
5WUA
 
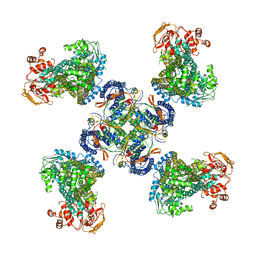 | | Structure of a Pancreatic ATP-sensitive Potassium Channel | | Descriptor: | ATP-sensitive inward rectifier potassium channel 11,superfolder GFP, SUR1 | | Authors: | Li, N, Wu, J.-X, Chen, L, Gao, N. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-25 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure of a Pancreatic ATP-Sensitive Potassium Channel
Cell, 168, 2017
|
|
1U4A
 
 | | Solution structure of human SUMO-3 C47S | | Descriptor: | Ubiquitin-like protein SMT3A | | Authors: | Ding, H, Xu, Y, Dai, H, Tang, Y, Wu, J, Shi, Y. | | Deposit date: | 2004-07-23 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human SUMO-3 C47S and Its Binding Surface for Ubc9
Biochemistry, 44, 2005
|
|
1RI6
 
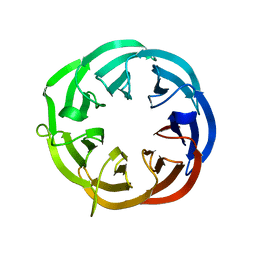 | | Structure of a putative isomerase from E. coli | | Descriptor: | putative isomerase ybhE | | Authors: | Lima, C.D, Kniewel, R, Solorzano, V, Wu, J, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-16 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a putative 7-bladed propeller isomerase
To be Published
|
|
8H8F
 
 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR (resting state) | | Descriptor: | Proton-activated chloride channel | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR (resting state)
To Be Published
|
|
