2JWE
 
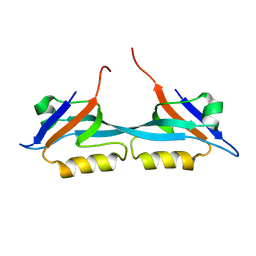 | | Solution structure of the second PDZ domain from human zonula occludens-1: A dimeric form with 3D domain swapping | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Ji, P, Wu, J.W, Zhang, J.H, Yang, Y.S, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2007-10-10 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second PDZ domain of Zonula Occludens 1
Proteins, 79, 2011
|
|
1DAV
 
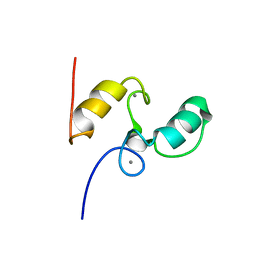 | | SOLUTION STRUCTURE OF THE TYPE I DOCKERIN DOMAIN FROM THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME (20 STRUCTURES) | | Descriptor: | CALCIUM ION, ENDOGLUCANASE SS | | Authors: | Lytle, B.L, Volkman, B.F, Westler, W.M, Heckman, M.P, Wu, J.H.D. | | Deposit date: | 1999-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain.
J.Mol.Biol., 307, 2001
|
|
2A5M
 
 | |
8TEC
 
 | |
5JI8
 
 | | Crystal structure of the BRD9 bromodomain and hit 1 | | Descriptor: | 2-amino-1,3-benzothiazole-6-carboxamide, Bromodomain-containing protein 9 | | Authors: | Wang, N, Li, F, Bao, H, Li, J, Wu, J, Ruan, K. | | Deposit date: | 2016-04-22 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | NMR Fragment Screening Hit Induces Plasticity of BRD7/9 Bromodomains
Chembiochem, 17, 2016
|
|
8TEE
 
 | |
1A7T
 
 | | METALLO-BETA-LACTAMASE WITH MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, METALLO-BETA-LACTAMASE, SODIUM ION, ... | | Authors: | Fitzgerald, P.M.D, Wu, J.K, Toney, J.H. | | Deposit date: | 1998-03-17 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unanticipated inhibition of the metallo-beta-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-A resolution.
Biochemistry, 37, 1998
|
|
1A8M
 
 | | TUMOR NECROSIS FACTOR ALPHA, R31D MUTANT | | Descriptor: | TUMOR NECROSIS FACTOR ALPHA | | Authors: | Reed, C, Fu, Z.-Q, Wu, J, Xue, Y.-N, Harrison, R.W, Chen, M.-J, Weber, I.T. | | Deposit date: | 1998-03-27 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TNF-alpha mutant R31D with greater affinity for receptor R1 compared with R2.
Protein Eng., 10, 1997
|
|
4X6R
 
 | | An Isoform-specific Myristylation Switch Targets RIIb PKA Holoenzymes to Membranes | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Zhang, P, Ye, F, Bastidas, A.C, Kornev, A.P, Ginsberg, M.H, Wu, J, Taylor, S.S. | | Deposit date: | 2014-12-09 | | Release date: | 2015-07-22 | | Last modified: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Isoform-Specific Myristylation Switch Targets Type II PKA Holoenzymes to Membranes.
Structure, 23, 2015
|
|
5KE4
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 6 nicotinic acetylcholine receptor in complex with 2-((5-(3,7-Diazabicyclo[3.3.1]nonan-3-yl)pyridin-3-yl)oxy)- N,N-dimethylethanamine (BPC) | | Descriptor: | 2-((5-(3,7-Diazabicyclo[3.3.1]nonan-3-yl)pyridin-3-yl)oxy)-N,N-dimethylethanamine, Soluble acetylcholine receptor | | Authors: | Bobango, J, Wu, J, Talley, I.T, Ralston, R, Sankaran, B, Talley, T.T. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 6 nicotinic acetylcholine receptor in complex with 2-((5-(3,7-Diazabicyclo[3.3.1]nonan-3-yl)pyridin-3-yl)oxy)-
N,N-dimethylethanamine (BPC)
To Be Published
|
|
8Q2M
 
 | |
5KZU
 
 | | Crystal structure of an acetylcholine binding protein from Aplysia californica (Ac-AChBP) in complex with click chemistry compound 9-[[1-[8-methyl-8-(2-phenylethyl)-8-azoniabicyclo[3.2.1]octan-3-yl]triazol-4-yl]methyl]carbazole | | Descriptor: | 9-[[1-[8-methyl-8-(2-phenylethyl)-8-azoniabicyclo[3.2.1]octan-3-yl]triazol-4-yl]methyl]carbazole, SULFATE ION, Soluble acetylcholine receptor, ... | | Authors: | Bobango, J, Wu, J, Talley, I.T, Sankaran, B, Talley, T.T. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) in complex with click chemistry compound 9-[[1-[8-methyl-8-(2-phenylethyl)-8-azoniabicyclo[3.2.1]octan-3-yl]triazol-4-yl]methyl]carbazole
To Be Published
|
|
1KN5
 
 | | SOLUTION STRUCTURE OF ARID DOMAIN OF ADR6 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-18 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
1KKX
 
 | | Solution structure of the DNA-binding domain of ADR6 | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-10 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
8IYJ
 
 | | Cryo-EM structure of the 48-nm repeat doublet microtubule from mouse sperm | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Zhou, L.N, Gui, M, Wu, J.P. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of sperm flagellar doublet microtubules expand the genetic spectrum of male infertility.
Cell, 186, 2023
|
|
8U4E
 
 | | Cryo-EM structure of long form insulin receptor (IR-B) with three IGF2 bound, asymmetric conformation. | | Descriptor: | Insulin receptor, Insulin-like growth factor II | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8U4B
 
 | | Cryo-EM structure of long form insulin receptor (IR-B) in the apo state | | Descriptor: | Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8U4C
 
 | | Cryo-EM structure of long form insulin receptor (IR-B) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin receptor, Insulin-like growth factor II | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
1L1Y
 
 | | The Crystal Structure and Catalytic Mechanism of Cellobiohydrolase CelS, the Major Enzymatic Component of the Clostridium thermocellum cellulosome | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, cellobiohydrolase | | Authors: | Guimaraes, B.G, Souchon, H, Lytle, B.L, Wu, J.H.D, Alzari, P.M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome.
J.Mol.Biol., 320, 2002
|
|
1L2A
 
 | | The Crystal Structure and Catalytic Mechanism of Cellobiohydrolase CelS, the Major Enzymatic Component of the Clostridium thermocellum cellulosome | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, cellobiohydrolase | | Authors: | Guimaraes, B.G, Souchon, H, Lytle, B.L, Wu, J.H.D, Alzari, P.M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome.
J.Mol.Biol., 320, 2002
|
|
2QUR
 
 | | Crystal Structure of F327A/K285P Mutant of cAMP-dependent Protein Kinase | | Descriptor: | 20-mer fragment from cAMP-dependent protein kinase inhibitor alpha, ADENOSINE-5'-DIPHOSPHATE, cAMP-dependent protein kinase, ... | | Authors: | Taylor, S.S, Yang, J, Wu, J. | | Deposit date: | 2007-08-06 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contribution of non-catalytic core residues to activity and regulation in protein kinase A.
J.Biol.Chem., 284, 2009
|
|
5JD0
 
 | | crystal structure of ARAP3 RhoGAP domain | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Bao, H, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2016-04-15 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Specific Recognition of RhoA by the Dual GTPase-activating Protein ARAP3
J.Biol.Chem., 291, 2016
|
|
4JCL
 
 | | Crystal structure of Alpha-CGT from Paenibacillus macerans at 1.7 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J, Wu, J, Li, J, Chen, J. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Alpha-Cgt from Paenibacillus Macerans at 1.7 Angstrom Resolution
To be Published
|
|
6W9E
 
 | | Crystal Structure of Human CDK9/cyclinT1 in complex with MC180295 | | Descriptor: | (4-amino-2-{[(1R,2R,4S)-bicyclo[2.2.1]heptan-2-yl]amino}-1,3-thiazol-5-yl)(2-nitrophenyl)methanone, (4-amino-2-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]amino}-1,3-thiazol-5-yl)(2-nitrophenyl)methanone, Cyclin-T1, ... | | Authors: | Zhang, P, Wu, J. | | Deposit date: | 2020-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Comparative Modeling of CDK9 Inhibitors to Explore Selectivity and Structure-Activity Relationships
To Be Published
|
|
1F9E
 
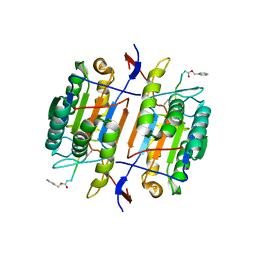 | | CASPASE-8 SPECIFICITY PROBED AT SUBSITE S4: CRYSTAL STRUCTURE OF THE CASPASE-8-Z-DEVD-CHO | | Descriptor: | (PHQ)DEVD, Caspase-8 subunit p10, Caspase-8 subunit p18 | | Authors: | Blanchard, H, Donepudi, M, Tschopp, M, Kodandapani, L, Wu, J.C, Grutter, M.G. | | Deposit date: | 2000-07-10 | | Release date: | 2001-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Caspase-8 specificity probed at subsite S(4): crystal structure of the caspase-8-Z-DEVD-cho complex.
J.Mol.Biol., 302, 2000
|
|
