7FA3
 
 | |
6JIF
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Pseudomonas sp. UW4 | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zheng, X, Guo, L, Li, D.F, Wu, B. | | Deposit date: | 2019-02-21 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of a highly active branched-chain amino acid aminotransferase from Pseudomonas sp. for efficient biosynthesis of chiral amino acids.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
5YUG
 
 | | AtVAL1 PHD-Like domain in the P31 space group | | Descriptor: | B3 domain-containing transcription repressor VAL1, GLYCEROL, ZINC ION | | Authors: | Zhang, M.M, Wu, B.X. | | Deposit date: | 2017-11-22 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | AtVAL1 PHD-Like domain in the P31 space group
To Be Published
|
|
7X5K
 
 | | Tir-dsDNA complex, the initial binding state | | Descriptor: | DNA (43-MER), Flax rust resistance protein | | Authors: | Tan, Y, Xu, C, Yu, D, Song, W, Wu, B, Schulze-Lefert, P, Chai, J. | | Deposit date: | 2022-03-04 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | TIR domains of plant immune receptors are 2',3'-cAMP/cGMP synthetases mediating cell death.
Cell, 185, 2022
|
|
6K7V
 
 | | Structure of NLRP1 CARD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 1 | | Authors: | Gong, Q, Xu, C, Zhang, J, Wu, B. | | Deposit date: | 2019-06-09 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8.
Nat Commun, 12, 2021
|
|
6K9F
 
 | | Structure of unknow protein 4 | | Descriptor: | Caspase recruitment domain-containing protein 8 | | Authors: | Gong, Q, Xu, C, Zhang, J, Boo, Z.Z, Wu, B. | | Deposit date: | 2019-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8.
Nat Commun, 12, 2021
|
|
7F8W
 
 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gq | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8V
 
 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gi | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
6K99
 
 | | Structure of ASC CARD filament | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD | | Authors: | Gong, Q, Xu, C, Zhang, J, Boo, Z.Z, Wu, B. | | Deposit date: | 2019-06-14 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8.
Nat Commun, 12, 2021
|
|
1J4I
 
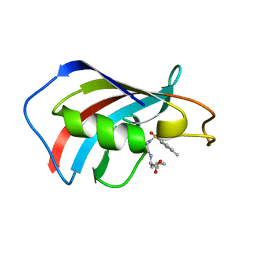 | | crystal structure analysis of the FKBP12 complexed with 000308 small molecule | | Descriptor: | 4-METHYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PENTANOIC ACID, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
1J4H
 
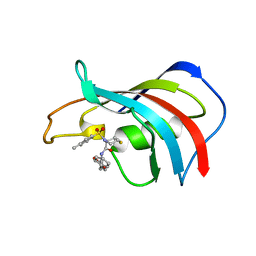 | | crystal structure analysis of the FKBP12 complexed with 000107 small molecule | | Descriptor: | 3-PHENYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PROPIONIC ACID ETHYL ESTER, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
5ZBH
 
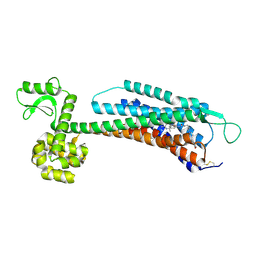 | | The Crystal Structure of Human Neuropeptide Y Y1 Receptor with BMS-193885 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme,Neuropeptide Y receptor type 1, dimethyl 4-{3-[({3-[4-(3-methoxyphenyl)piperidin-1-yl]propyl}carbamoyl)amino]phenyl}-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-11 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
6K8J
 
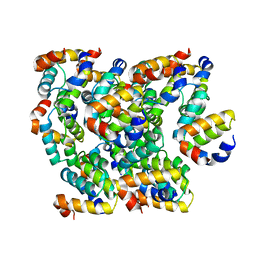 | | Structure of NLRC4 CARD filament | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Gong, Q, Xu, C, Zhang, J, Boo, Z.Z, Wu, B. | | Deposit date: | 2019-06-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8.
Nat Commun, 12, 2021
|
|
5ZBQ
 
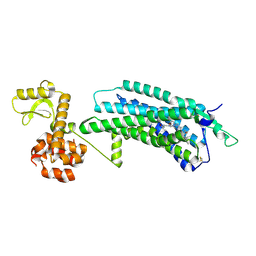 | | The Crystal Structure of human neuropeptide Y Y1 receptor with UR-MK299 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme, N~2~-(diphenylacetyl)-N-[(4-hydroxyphenyl)methyl]-N~5~-(N'-{[2-(propanoylamino)ethyl]carbamoyl}carbamimidoyl)-D-ornithinamide | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
6KY5
 
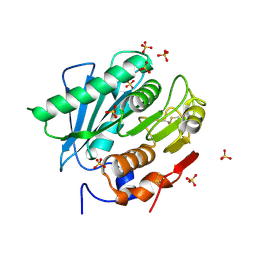 | | Crystal structure of a hydrolase mutant | | Descriptor: | PET hydrolase, SULFATE ION | | Authors: | Cui, Y.L, Chen, Y.C, Liu, X.Y, Dong, S.J, Han, J, Xiang, H, Chen, Q, Liu, H.Y, Han, X, Liu, W.D, Tang, S.Y, Wu, B. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Computational redesign of PETase for plasticbiodegradation by GRAPE strategy.
Biorxiv, 2020
|
|
5YQZ
 
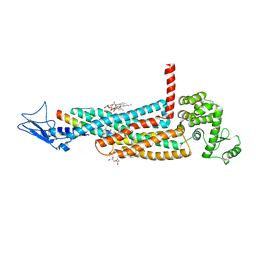 | | Structure of the glucagon receptor in complex with a glucagon analogue | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon analogue, ... | | Authors: | Zhang, H, Qiao, A, Yang, L, VAN EPS, N, Frederiksen, K, Yang, D, Dai, A, Cai, X, Zhang, H, Yi, C, Can, C, He, L, Yang, H, Lau, J, Ernst, O, Hanson, M, Stevens, R, Wang, M, Seedtz-Runge, S, Jiang, H, Zhao, Q, Wu, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the glucagon receptor in complex with a glucagon analogue.
Nature, 553, 2018
|
|
5ZKQ
 
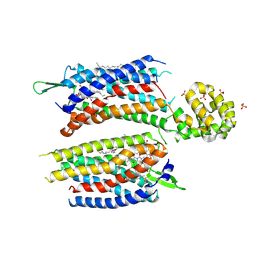 | | Crystal structure of the human platelet-activating factor receptor in complex with ABT-491 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-ethynyl-3-{3-fluoro-4-[(2-methyl-1H-imidazo[4,5-c]pyridin-1-yl)methyl]benzene-1-carbonyl}-N,N-dimethyl-1H-indole-1-carboxamide, Platelet-activating factor receptor,Endolysin,Endolysin,Platelet-activating factor receptor, ... | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3MKV
 
 | | Crystal structure of amidohydrolase eaj56179 | | Descriptor: | CARBONATE ION, GLYCEROL, PUTATIVE AMIDOHYDROLASE, ... | | Authors: | Patskovsky, Y, Bonanno, J, Ozyurt, S, Sauder, J.M, Freeman, J, Wu, B, Smith, D, Bain, K, Rodgers, L, Wasserman, S.R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-15 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3NEK
 
 | | Crystal structure of a nitrogen repressor-like protein MJ0159 from Methanococcus jannaschii | | Descriptor: | GLYCEROL, nitrogen repressor-like protein MJ0159 | | Authors: | Bonanno, J.B, Patskovsky, Y, Malashkevich, V, Ozyurt, S, Dickey, M, Wu, B, Maletic, M, Rodgers, L, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-06-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural underpinnings of nitrogen regulation by the prototypical nitrogen-responsive transcriptional factor NrpR.
Structure, 18, 2010
|
|
2P4U
 
 | | Crystal structure of acid phosphatase 1 (Acp1) from Mus musculus | | Descriptor: | Acid phosphatase 1, PHOSPHATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-13 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
