6GUH
 
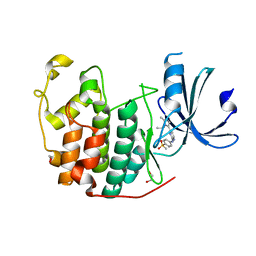 | | CDK2 in complex with AZD5438 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GU3
 
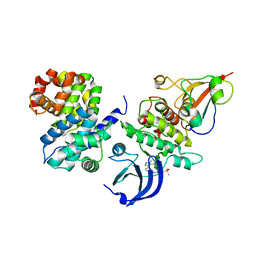 | | CDK1/CyclinB/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GU2
 
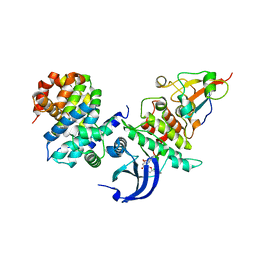 | | CDK1/CyclinB/Cks2 in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GUK
 
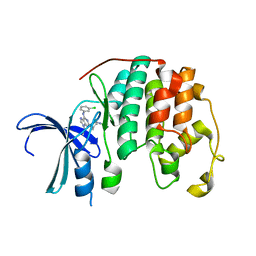 | | CDK2 in complex with CGP74514A | | Descriptor: | Cyclin-dependent kinase 2, ~{N}2-[(1~{R},2~{S})-2-azanylcyclohexyl]-~{N}6-(3-chlorophenyl)-9-ethyl-purine-2,6-diamine | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
1JTW
 
 | |
1TFN
 
 | |
1CP2
 
 | | NITROGENASE IRON PROTEIN FROM CLOSTRIDIUM PASTEURIANUM | | Descriptor: | IRON/SULFUR CLUSTER, NITROGENASE IRON PROTEIN | | Authors: | Schlessman, J.L, Woo, D, Joshua-Tor, L, Howard, J.B, Rees, D.C. | | Deposit date: | 1998-05-11 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum.
J.Mol.Biol., 280, 1998
|
|
2NIP
 
 | | NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | IRON/SULFUR CLUSTER, NITROGENASE IRON PROTEIN | | Authors: | Komiya, H, Georgiadis, M.M, Chakrabarti, P, Woo, D, Kornuc, J.J, Rees, D.C. | | Deposit date: | 1998-05-11 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum.
J.Mol.Biol., 280, 1998
|
|
1NIP
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF THE NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Komiya, H, Georgiadis, M.M, Chakrabarti, P, Woo, D, Kornuc, J.J, Rees, D.C. | | Deposit date: | 1992-09-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii.
Science, 257, 1992
|
|
1MIO
 
 | | X-RAY CRYSTAL STRUCTURE OF THE NITROGENASE MOLYBDENUM-IRON PROTEIN FROM CLOSTRIDIUM PASTEURIANUM AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE-MO-S CLUSTER, ... | | Authors: | Kim, J, Woo, D, Rees, D.C. | | Deposit date: | 1993-03-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of the nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 3.0-A resolution.
Biochemistry, 32, 1993
|
|
4PN9
 
 | | A de novo designed hexameric coiled coil CC-Hex2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CC-Hex2 | | Authors: | Wood, C.W, Burton, A.J, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
4PNA
 
 | | A de novo designed heptameric coiled coil CC-Hept | | Descriptor: | CC-Hept, GLYCEROL | | Authors: | Burton, A.J, Wood, C.W, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
4PND
 
 | | A de novo designed pentameric coiled coil CC-Pent_Variant | | Descriptor: | CC-Pent_Variant | | Authors: | Wood, C.W, Burton, A.J, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
4PNB
 
 | | A de novo designed hexameric coiled coil CC-Hex3. | | Descriptor: | CC-Hex3 | | Authors: | Wood, C.W, Burton, A.J, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
6XY1
 
 | |
1EYM
 
 | | FK506 BINDING PROTEIN MUTANT, HOMODIMERIC COMPLEX | | Descriptor: | FK506 BINDING PROTEIN | | Authors: | Rollins, C.T, Rivera, V.M, Woolfson, D.N, Keenan, T, Hatada, M, Adams, S.E, Andrade, L.J, Yaeger, D, van Schravendijk, M.R, Holt, D.A, Gilman, M, Clackson, T. | | Deposit date: | 2000-05-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ligand-reversible dimerization system for controlling protein-protein interactions.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6XY0
 
 | |
5A0E
 
 | | Crystal structure of cyclophilin D in complex with CsA analogue, JW47. | | Descriptor: | JW47, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE F, MITOCHONDRIAL | | Authors: | Warne, J, Pryce, G, Hill, J, Shi, X, Lenneras, F, Puentes, F, Kip, M, Hilditch, L, Walker, P, Simone, M, Chan, A.W.E, Towers, G, Coker, A.R, Duchen, M, Szabadkai, G, Baker, D, Selwood, D.L. | | Deposit date: | 2015-04-19 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selective Inhibition of the Mitochondrial Permeability Transition Pore Protects Against Neuro-Degeneration in Experimental Multiple Sclerosis.
J.Biol.Chem., 291, 2016
|
|
6EIK
 
 | | A de novo designed heptameric coiled coil CC-Hept-I24E | | Descriptor: | CC-Hept-I24E, FORMIC ACID, GLYCEROL | | Authors: | Rhys, G.G, Burton, A.J, Dawson, W.M, Thomas, F, Woolfson, D.N. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | De Novo-Designed alpha-Helical Barrels as Receptors for Small Molecules.
ACS Synth Biol, 7, 2018
|
|
8A09
 
 | | A hexameric barrel state of a de novo coiled-coil assembly: CC-Type2-(QgLaId)4 | | Descriptor: | A hexameric barrel state of a de novo coiled-coil assembly: CC-Type2-(QgLaId)4, GLYCEROL | | Authors: | Martin, F.J.O, Rhys, G.G, Dawson, W.M, Woolfson, D.N. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Differential sensing with arrays of de novo designed peptide assemblies.
Nat Commun, 14, 2023
|
|
6E3Y
 
 | | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor | | Descriptor: | Calcitonin gene-related peptide 1, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Deganutti, G, Glukhova, A, Koole, C, Peat, T.S, Radjainia, M, Plitzko, J.M, Baumeister, W, Miller, L.J, Hay, D.L, Christopoulos, A, Reynolds, C.A, Wootten, D, Sexton, P.M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor.
Nature, 561, 2018
|
|
2G0L
 
 | | Solution Structure of Neocarzinostatin Apo-Protein with bound Flavone | | Descriptor: | 2-PHENYL-4H-CHROMEN-4-ONE, NEOCARZINOSTATIN | | Authors: | Muskett, F.W, Stoneman, R.G, Caddick, S, Woolfson, D.N. | | Deposit date: | 2006-02-13 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Synthetic Ligands for Apo-Neocarzinostatin
J.Am.Chem.Soc., 128, 2006
|
|
6D8C
 
 | | Cryo-EM structure of FLNaABD E254K bound to phalloidin-stabilized F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Iwamoto, D.V, Huehn, A.R, Simon, B, Huet-Calderwood, C, Baldassarre, M, Sindelar, C.V, Calderwood, D.A. | | Deposit date: | 2018-04-26 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of the filamin A actin-binding domain interaction with F-actin.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6Q5N
 
 | | Crystal structure of a CC-Hex mutant that forms a parallel six-helix coiled coil CC-Hex*-L24Nle | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CC-Hex*-L24Nle, GLYCEROL | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5R
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-LL-KgEb | | Descriptor: | CC-Hex*-LL-KgEb, GLYCEROL | | Authors: | Beesley, J.L, Rhys, G.G, Wood, C.W, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
