2WSM
 
 | |
5K35
 
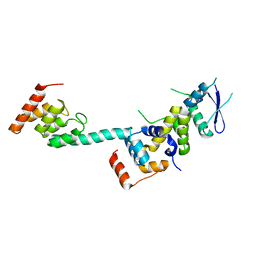 | | Structure of the Legionella effector, AnkB, in complex with human Skp1 | | Descriptor: | Ankyrin-repeat protein B, S-phase kinase-associated protein 1 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
5K34
 
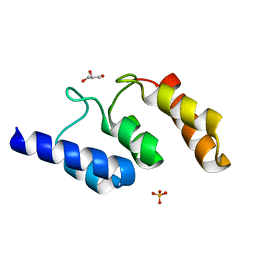 | | Structure of the ankyrin domain of AnkB from Legionella Pneumophila | | Descriptor: | Ankyrin-repeat protein B, GLYCEROL, SULFATE ION | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
1AB7
 
 | |
5BU2
 
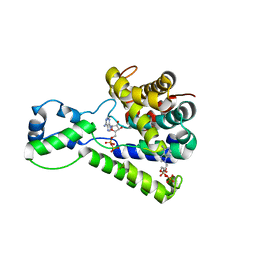 | | Structure of the C-terminal domain of lpg1496 from Legionella pneumophila in complex with nucleotide | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, alpha-D-ribofuranose, ... | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BTZ
 
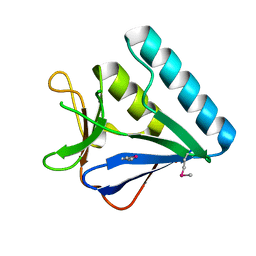 | | Structure of the middle domain of lpg1496 from Legionella pneumophila in P212121 space group | | Descriptor: | lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BTX
 
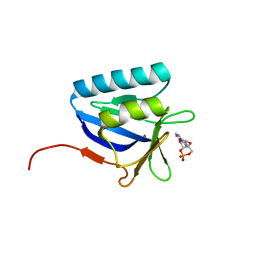 | | Structure of the N-terminal domain of lpg1496 from Legionella pneumophila in complex with nucleotide | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BU0
 
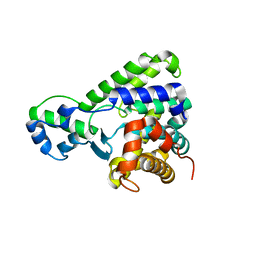 | | Structure of the C-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BTW
 
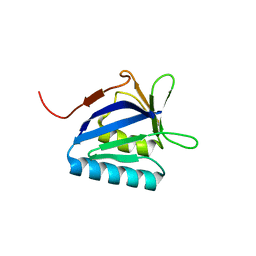 | | Structure of the N-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BU1
 
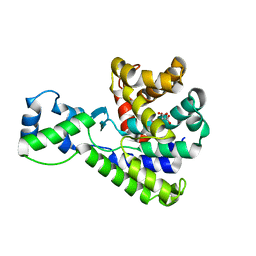 | | Structure of the truncated C-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | LPG1496, MALONATE ION | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5XIV
 
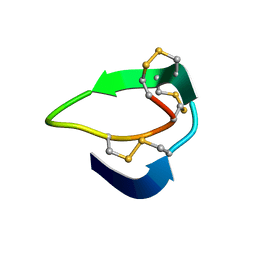 | | Beta-Ginkgotides: Hyperdisulfide-constrained peptides from Ginkgo biloba | | Descriptor: | beta-ginkgotide, beta-gB1 | | Authors: | Wong, K.H, Tan, W.L, Xiao, T, Tam, J.P. | | Deposit date: | 2017-04-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | beta-Ginkgotides: Hyperdisulfide-constrained peptides from Ginkgo biloba.
Sci Rep, 7, 2017
|
|
1Z9I
 
 | |
1QHM
 
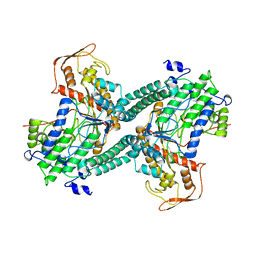 | | ESCHERICHIA COLI PYRUVATE FORMATE LYASE LARGE DOMAIN | | Descriptor: | PYRUVATE FORMATE-LYASE | | Authors: | Leppanen, V.-M, Merckel, M.C, Ollis, D.L, Wong, K.K, Kozarich, J.W, Goldman, A. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyruvate formate lyase is structurally homologous to type I ribonucleotide reductase.
Structure Fold.Des., 7, 1999
|
|
1H7M
 
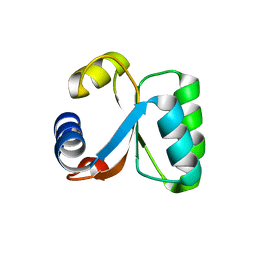 | | Ribosomal Protein L30e from Thermococcus celer | | Descriptor: | 50S RIBOSOMAL PROTEIN L30E | | Authors: | Chen, Y.W, Wong, K.B. | | Deposit date: | 2001-07-09 | | Release date: | 2003-04-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Ribosomal Protein L30E from the Extreme Thermophile Thermocccus Celer: Thermal Stability and RNA Binding
Biochemistry, 42, 2003
|
|
1GO1
 
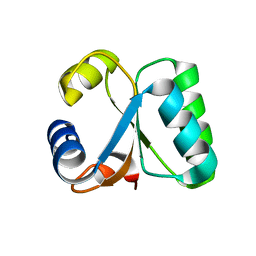 | |
7BVW
 
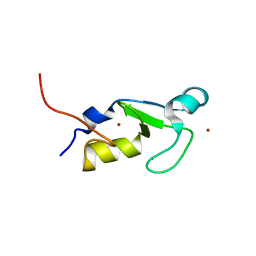 | | Crystal structure of the RING-H2 domain of Arabidopsis RMR1 | | Descriptor: | AT5G66160 protein, SODIUM ION, ZINC ION | | Authors: | Chen, S, Wong, K.B. | | Deposit date: | 2020-04-12 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The RING-finger of AtRMR1 (Arabidopsis receptor-homology-transmembrane-RING-H2 sorting receptor 1) is an E3 ligase that mediate its trafficking
To Be Published
|
|
6UAN
 
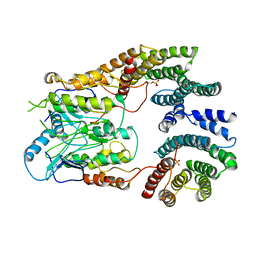 | | B-Raf:14-3-3 complex | | Descriptor: | 14-3-3 zeta, Serine/threonine-protein kinase B-raf | | Authors: | Kondo, Y, Ognjenovic, J, Banerjee, S, Karandur, D, Merk, A, Kulhanek, K, Wong, K, Roose, J.P, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Science, 366, 2019
|
|
5WD8
 
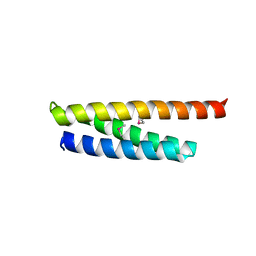 | |
5WD9
 
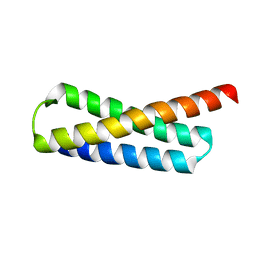 | |
2PQI
 
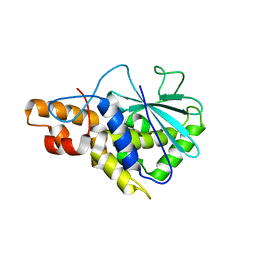 | | Crystal structure of active ribosome inactivating protein from maize (b-32) | | Descriptor: | Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Wong, Y.T, Young, J.A, Cha, S.S, Sze, K.H, Au, S.W.N, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
2PQG
 
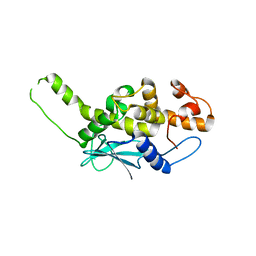 | | Crystal structure of inactive ribosome inactivating protein from maize (b-32) | | Descriptor: | Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Wong, Y.T, Young, J.A, Cha, S.S, Sze, K.H, Au, S.W.N, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
8HC1
 
 | | CryoEM structure of Helicobacter pylori UreFD/urease complex | | Descriptor: | Urease accessory protein UreF, Urease accessory protein UreH, Urease subunit alpha, ... | | Authors: | Nim, Y.S, Fong, I.Y.H, Deme, J, Tsang, K.L, Caesar, J, Johnson, S, Wong, K.B, Lea, S.M. | | Deposit date: | 2022-11-01 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Delivering a toxic metal to the active site of urease.
Sci Adv, 9, 2023
|
|
8HCN
 
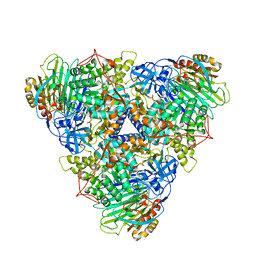 | | CryoEM Structure of Klebsiella pneumoniae UreD/urease complex | | Descriptor: | Urease accessory protein UreD, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Nim, Y.S, Fong, I.Y.H, Deme, J, Tsang, K.L, Caesar, J, Johnson, S, Wong, K.B, Lea, S.M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Delivering a toxic metal to the active site of urease.
Sci Adv, 9, 2023
|
|
8KD5
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
