1XJH
 
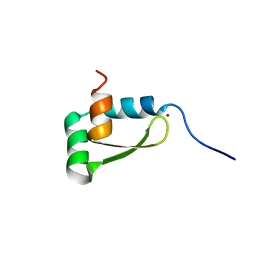 | | NMR structure of the redox switch domain of the E. coli Hsp33 | | Descriptor: | 33 kDa chaperonin, ZINC ION | | Authors: | Won, H.S, Low, L.Y, De Guzman, R.N, Martinez-Yamout, M.A, Jakob, U, Dyson, H.J. | | Deposit date: | 2004-09-23 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Zinc-dependent Redox Switch Domain of the Chaperone Hsp33 has a Novel Fold
J.Mol.Biol., 341, 2004
|
|
8QMO
 
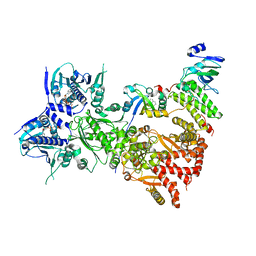 | | Cryo-EM structure of the benzo[a]pyrene-bound Hsp90-XAP2-AHR complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Kwong, H.S, Grandvuillemin, L, Sirounian, S, Ancelin, A, Lai-Kee-Him, J, Carivenc, C, Lancey, C, Ragan, T.J, Hesketh, E.L, Bourguet, W, Gruszczyk, J. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Insights into the Activation of Human Aryl Hydrocarbon Receptor by the Environmental Contaminant Benzo[a]pyrene and Structurally Related Compounds.
J.Mol.Biol., 436, 2024
|
|
7X89
 
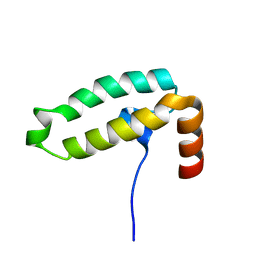 | | Tid1 | | Descriptor: | DnaJ homolog subfamily A member 3, mitochondrial | | Authors: | Jang, J, Lee, S.H, Kang, D.H, Sim, D.W, Jo, K.S, Ryu, H, Kim, E.H, Ryu, K.S, Lee, J.H, Kim, J.H, Won, H.S. | | Deposit date: | 2022-03-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the J-domain and GF-motif of the mitochondrial Hsp40, Tid1
To Be Published
|
|
5J4G
 
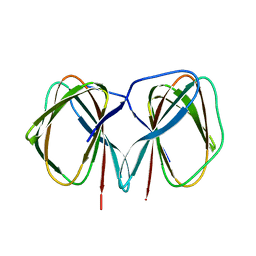 | | Crystal structure of the C-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | Descriptor: | Uncharacterized protein | | Authors: | Sim, D.W, Lee, W.C, Kim, H.Y, Kim, J.H, Won, H.S. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
6IWS
 
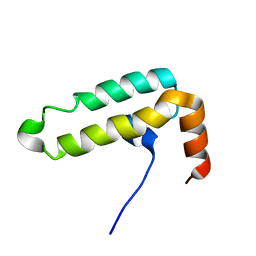 | | Solution structure of the J-domain of Tid1, a Mitochondrial Hsp40/DnaJ Protein | | Descriptor: | DnaJ homolog subfamily A member 3, mitochondrial | | Authors: | Sim, D.W, Jo, K.S, Won, H.S, Kim, J.H. | | Deposit date: | 2018-12-06 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the J-domain of Tid1, a Mitochondrial Hsp40/DnaJ Protein
To Be Published
|
|
2MC4
 
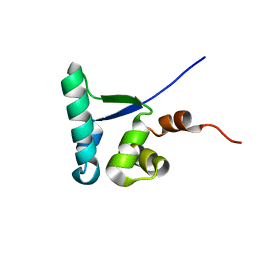 | |
5I76
 
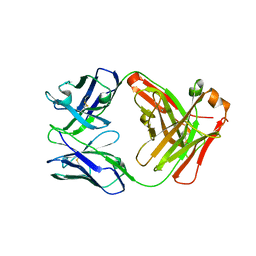 | | Crystal structure of FM318, a recombinant Fab adopted from cetuximab | | Descriptor: | FM318_heavy_cahin, FM318_light_chain | | Authors: | Sim, D.W, Kim, J.H, Seok, S.H, Seo, M.D, Kim, Y.P, Won, H.S. | | Deposit date: | 2016-02-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Bacterial production and structure-functional validation of a recombinant antigen-binding fragment (Fab) of an anti-cancer therapeutic antibody targeting epidermal growth factor receptor.
Appl.Microbiol.Biotechnol., 100, 2016
|
|
5GZ0
 
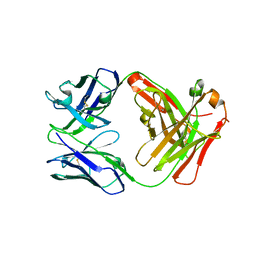 | | Crystal structure of FM329, a recombinant Fab adopted from cetuximab | | Descriptor: | FM329 heavy chain, FM329 light chain | | Authors: | Sim, D.W, Kim, J.H, Kim, Y.P, Won, H.S. | | Deposit date: | 2016-09-26 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of FM329, a recombinant Fab adopted from cetuximab
To Be Published
|
|
7ZUB
 
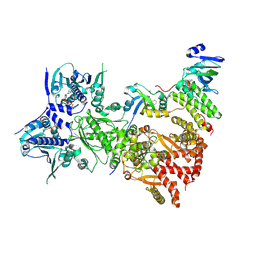 | | Cryo-EM structure of the indirubin-bound Hsp90-XAP2-AHR complex | | Descriptor: | (3~{Z})-3-(3-oxidanylidene-1~{H}-indol-2-ylidene)-1~{H}-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, ... | | Authors: | Gruszczyk, J, Savva, C.G, Lai-Kee-Him, J, Bous, J, Ancelin, A, Kwong, H.S, Grandvuillemin, L, Bourguet, W. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the agonist-bound Hsp90-XAP2-AHR cytosolic complex.
Nat Commun, 13, 2022
|
|
