8OKF
 
 | | WD repeat containing protein 5 (WDR5)- PER2 peptide | | Descriptor: | CHLORIDE ION, Glutathione S-transferase class-mu 26 kDa isozyme,WD repeat domain 5, PENTAETHYLENE GLYCOL, ... | | Authors: | Wolf, E, Boergel, A. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A structural competition involving WDR5 times circadian oscillations
To Be Published
|
|
8OK1
 
 | |
1KUY
 
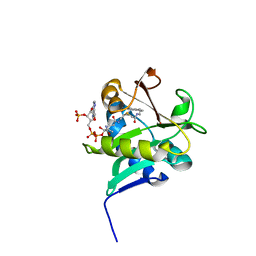 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-ACETYL TRYPTAMINE, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
1KUV
 
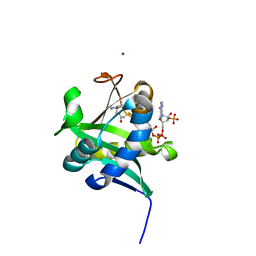 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-ACETYL 5-BROMOTRYPTAMINE, MAGNESIUM ION, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
1KUX
 
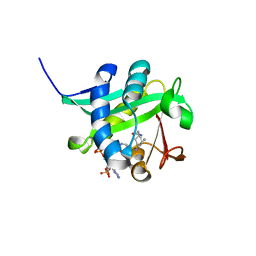 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-TRIMETHYLENE-ACETYL-TRYPTAMINE, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
1BO4
 
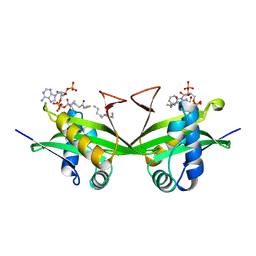 | | CRYSTAL STRUCTURE OF A GCN5-RELATED N-ACETYLTRANSFERASE: SERRATIA MARESCENS AMINOGLYCOSIDE 3-N-ACETYLTRANSFERASE | | Descriptor: | COENZYME A, PROTEIN (SERRATIA MARCESCENS AMINOGLYCOSIDE-3-N-ACETYLTRANSFERASE), SPERMIDINE | | Authors: | Wolf, E, Vassilev, A, Makino, Y, Sali, A, Nakatani, Y, Burley, S.K. | | Deposit date: | 1998-08-08 | | Release date: | 1998-10-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase.
Cell(Cambridge,Mass.), 94, 1998
|
|
4DJ3
 
 | |
2YMY
 
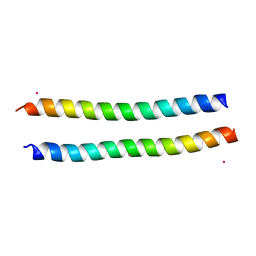 | | Structure of the murine Nore1-Sarah domain | | Descriptor: | CADMIUM ION, RAS ASSOCIATION DOMAIN-CONTAINING PROTEIN 5 | | Authors: | Makbul, C, Schwarz, D, Aruxandei, D.C, Wolf, E, Hofmann, E, Herrmann, C. | | Deposit date: | 2012-10-10 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and Thermodynamic Characterization of Nore1-Sarah: A Small, Helical Module Important in Signal Transduction Networks.
Biochemistry, 52, 2013
|
|
3GEC
 
 | |
3GDI
 
 | |
4CT0
 
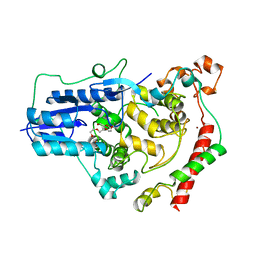 | | Crystal Structure of Mouse Cryptochrome1 in Complex with Period2 | | Descriptor: | CHLORIDE ION, CRYPTOCHROME-1, HEXAETHYLENE GLYCOL, ... | | Authors: | Schmalen, I, Rajan Prabu, J, Benda, C, Wolf, E. | | Deposit date: | 2014-03-11 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Interaction of Circadian Clock Proteins Cry1 and Per2 is Modulated by Zinc Binding and Disulfide Bond Formation.
Cell(Cambridge,Mass.), 157, 2014
|
|
4DJ2
 
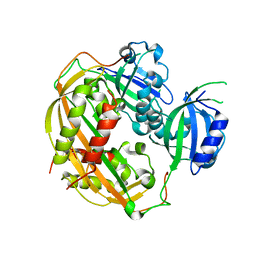 | | Unwinding the Differences of the Mammalian PERIOD Clock Proteins from Crystal Structure to Cellular Function | | Descriptor: | Period circadian protein homolog 1 | | Authors: | Kucera, N, Schmalen, I, Hennig, S, Oellinger, R, Strauss, H.M, Grudziecki, A, Wieczorek, C, Kramer, A, Wolf, E. | | Deposit date: | 2012-02-01 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Unwinding the differences of the mammalian PERIOD clock proteins from crystal structure to cellular function.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1B9G
 
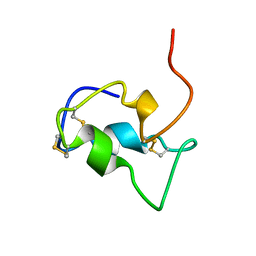 | | INSULIN-LIKE-GROWTH-FACTOR-1 | | Descriptor: | PROTEIN (GROWTH FACTOR IGF-1) | | Authors: | De Wolf, E, Gill, R, Geddes, S, Pitts, J, Wollmer, A, Grotzinger, J. | | Deposit date: | 1999-02-11 | | Release date: | 1999-02-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a mini IGF-1.
Protein Sci., 5, 1996
|
|
1HE1
 
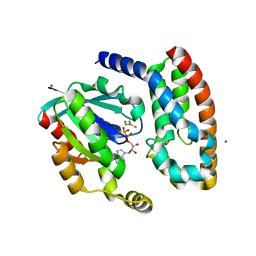 | | Crystal structure of the complex between the GAP domain of the Pseudomonas aeruginosa ExoS toxin and human Rac | | Descriptor: | ALUMINUM FLUORIDE, EXOENZYME S, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wurtele, M, Wolf, E, Pederson, K.J, Buchwald, G, Ahmadian, M.R, Barbieri, J.T, Wittinghofer, A. | | Deposit date: | 2000-11-18 | | Release date: | 2001-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How the Pseudomonas Aeruginosa Exos Toxin Downregulates Rac
Nat.Struct.Biol., 8, 2001
|
|
1HE9
 
 | | Crystal structure of the GAP domain of the Pseudomonas aeruginosa ExoS toxin | | Descriptor: | EXOENZYME S | | Authors: | Wurtele, M, Renault, L, Barbieri, J.T, Wittinghofer, A, Wolf, E. | | Deposit date: | 2000-11-21 | | Release date: | 2001-03-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Exos Gtpase Activating Domain
FEBS Lett., 491, 2001
|
|
1TPZ
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQ2
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQD
 
 | | Crystal structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, interferon-inducible GTPase | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1O7F
 
 | | CRYSTAL STRUCTURE OF THE REGULATORY DOMAIN OF EPAC2 | | Descriptor: | CAMP-DEPENDENT RAP1 GUANINE-NUCLEOTIDE EXCHANGE FACTOR | | Authors: | Rehmann, H, Prakash, B, Wolf, E, Rueppel, A, De Rooij, J, Bos, J.L, Wittinghofer, A. | | Deposit date: | 2002-11-04 | | Release date: | 2002-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Regulation of the Camp-Binding Domains of Epac2
Nat.Struct.Biol., 10, 2002
|
|
1TQ4
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, interferon-inducible GTPase | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1WA9
 
 | | Crystal Structure of the PAS repeat region of the Drosophila clock protein PERIOD | | Descriptor: | PERIOD CIRCADIAN PROTEIN | | Authors: | Yildiz, O, Doi, M, Yujnovsky, I, Cardone, L, Berndt, A, Hennig, S, Schulze, S, Urbanke, C, Sassone-Corsi, P, Wolf, E. | | Deposit date: | 2004-10-25 | | Release date: | 2005-01-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure and Interactions of the Pas Repeat Region of the Drosophila Clock Protein Period
Mol.Cell, 17, 2005
|
|
1TQ6
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, interferon-inducible GTPase | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1USD
 
 | | human VASP tetramerisation domain L352M | | Descriptor: | VASODILATOR-STIMULATED PHOSPHOPROTEIN | | Authors: | Kuhnel, K, Jarchau, T, Wolf, E, Schlichting, I, Walter, U, Wittinghofer, A, Strelkov, S.V. | | Deposit date: | 2003-11-21 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Vasp Tetramerization Domain is a Right-Handed Coiled Coil Based on a 15-Residue Repeat
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1USE
 
 | | human VASP tetramerisation domain | | Descriptor: | VASODILATOR-STIMULATED PHOSPHOPROTEIN | | Authors: | Kuhnel, K, Jarchau, T, Wolf, E, Schlichting, I, Walter, U, Wittinghofer, A, Strelkov, S.V. | | Deposit date: | 2003-11-21 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Vasp Tetramerization Domain is a Right-Handed Coiled Coil Based on a 15-Residue Repeat
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7Q2J
 
 | | Quaternary Complex of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC Homer | | Descriptor: | Elongin-B, Elongin-C, N-[5-[4-[[5-[[(2S)-3,3-dimethyl-1-[(2S,4R)-2-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methylcarbamoyl]-4-oxidanyl-pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]amino]-5-oxidanylidene-pentyl]carbamoyl]phenyl]-2-(4-methylpiperazin-1-yl)phenyl]-6-oxidanylidene-4-(trifluoromethyl)-1H-pyridine-3-carboxamide, ... | | Authors: | Kraemer, A, Doelle, A, Schwalm, M.P, Adhikari, B, Wolf, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-10-25 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
