5H84
 
 | | Human Gcn5 bound to propionyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, Histone acetyltransferase KAT2A, ISOPROPYL ALCOHOL, ... | | Authors: | Wolberger, C, Ringel, A.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for acyl-group discrimination by human Gcn5L2.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5H86
 
 | | Human Gcn5 bound to butyryl-CoA | | Descriptor: | 1,2-ETHANEDIOL, Butyryl Coenzyme A, Histone acetyltransferase KAT2A | | Authors: | Wolberger, C, Ringel, A.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for acyl-group discrimination by human Gcn5L2.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4WA6
 
 | |
4W4U
 
 | |
1APL
 
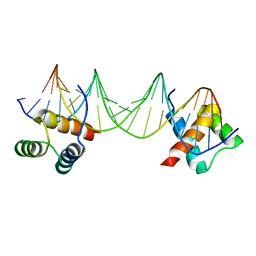 | | CRYSTAL STRUCTURE OF A MAT-ALPHA2 HOMEODOMAIN-OPERATOR COMPLEX SUGGESTS A GENERAL MODEL FOR HOMEODOMAIN-DNA INTERACTIONS | | Descriptor: | DNA (5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*C P*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MAT-ALPHA2 HOMEODOMAIN) | | Authors: | Wolberger, C, Vershon, A.K, Liu, B, Johnson, A.D, Pabo, C.O. | | Deposit date: | 1993-10-04 | | Release date: | 1993-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions.
Cell(Cambridge,Mass.), 67, 1991
|
|
3PDH
 
 | |
2GMI
 
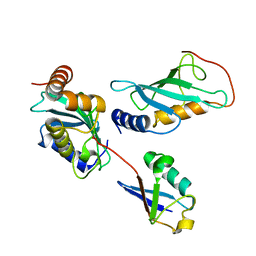 | | Mms2/Ubc13~Ubiquitin | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 13, Ubiquitin-conjugating enzyme variant MMS2 | | Authors: | Wolberger, C, Eddins, M.J, Carlile, C.M, Gomez, K.G, Pickart, C.M. | | Deposit date: | 2006-04-06 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3D4B
 
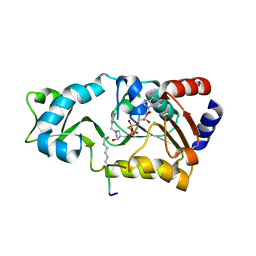 | | Crystal structure of Sir2Tm in complex with Acetyl p53 peptide and DADMe-NAD+ | | Descriptor: | 5'-O-[(R)-{[(R)-{[(3R,4R)-1-(3-carbamoylbenzyl)-4-hydroxypyrrolidin-3-yl]methoxy}(hydroxy)phosphoryl]methyl}(hydroxy)phosphoryl]adenosine, Acetyl P53 peptide, NAD-dependent deacetylase, ... | | Authors: | Hawse, W.F, Hoff, K.G, Fatkins, D, Daines, A, Zubkova, O.V, Schramm, V.L, Zheng, W, Wolberger, C. | | Deposit date: | 2008-05-14 | | Release date: | 2008-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into intermediate steps in the Sir2 deacetylation reaction.
Structure, 16, 2008
|
|
4FK5
 
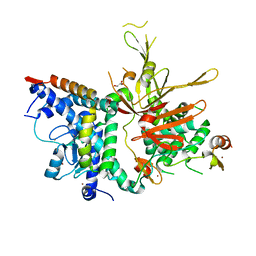 | | Structure of the SAGA Ubp8(S144N)/Sgf11/Sus1/Sgf73 DUB module | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein SUS1, ... | | Authors: | Samara, N.L, Ringel, A.E, Wolberger, C. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | A Role for Intersubunit Interactions in Maintaining SAGA Deubiquitinating Module Structure and Activity.
Structure, 20, 2012
|
|
4FJC
 
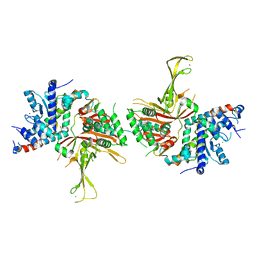 | | Structure of the SAGA Ubp8/Sgf11(1-72, Delta-ZnF)/Sus1/Sgf73 DUB module | | Descriptor: | GLYCEROL, Protein SUS1, SAGA-associated factor 11, ... | | Authors: | Samara, N.L, Ringel, A.E, Wolberger, C. | | Deposit date: | 2012-06-11 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.826 Å) | | Cite: | A Role for Intersubunit Interactions in Maintaining SAGA Deubiquitinating Module Structure and Activity.
Structure, 20, 2012
|
|
1MDM
 
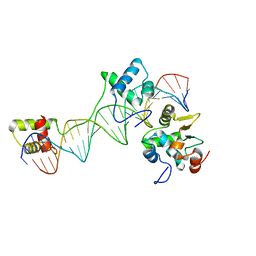 | | INHIBITED FRAGMENT OF ETS-1 AND PAIRED DOMAIN OF PAX5 BOUND TO DNA | | Descriptor: | C-ETS-1 PROTEIN, PAIRED BOX PROTEIN PAX-5, PAX5/ETS BINDING SITE ON THE MB-1 PROMOTER | | Authors: | Garvie, C.W, Pufall, M.A, Graves, B.J, Wolberger, C. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | STRUCTURAL ANALYSIS OF THE AUTOINHIBITION OF ETS-1 AND ITS ROLE IN PROTEIN PARTNERSHIPS
J.Biol.Chem., 277, 2002
|
|
8G6H
 
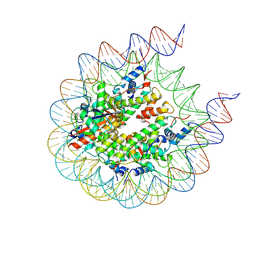 | |
7UD5
 
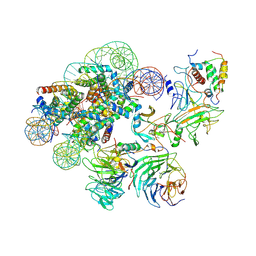 | | Complex between MLL1-WRAD and an H2B-ubiquitinated nucleosome | | Descriptor: | 601 DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Niklas, H.A, Rahman, S, Worden, E.J, Wolberger, C. | | Deposit date: | 2022-03-18 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Multistate structures of the MLL1-WRAD complex bound to H2B-ubiquitinated nucleosome.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1MD0
 
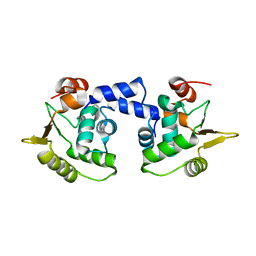 | |
8DU4
 
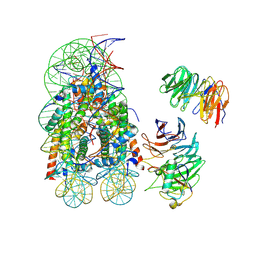 | | Complex between RbBP5-WDR5 and an H2B-ubiquitinated nucleosome | | Descriptor: | 601 DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Niklas, H.A, Rahman, S, Worden, E.J, Wolberger, C. | | Deposit date: | 2022-07-26 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Multistate structures of the MLL1-WRAD complex bound to H2B-ubiquitinated nucleosome.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1MH3
 
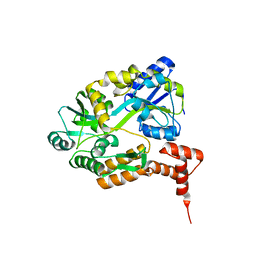 | | maltose binding-a1 homeodomain protein chimera, crystal form I | | Descriptor: | maltose binding-a1 homeodomain protein chimera | | Authors: | Ke, A, Wolberger, C. | | Deposit date: | 2002-08-19 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into binding cooperativity of MATa1/MATalpha2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera
Protein Sci., 12, 2003
|
|
1MH4
 
 | | maltose binding-a1 homeodomain protein chimera, crystal form II | | Descriptor: | maltose binding-a1 homeodomain protein chimera | | Authors: | Ke, A, Wolberger, C. | | Deposit date: | 2002-08-19 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into binding cooperativity of MATa1/MATalpha2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera
Protein Sci., 12, 2003
|
|
4TWI
 
 | | The structure of Sir2Af1 bound to a succinylated histone peptide | | Descriptor: | GLYCEROL, NAD-dependent protein deacylase 1, Succinylated H4 Peptide (aa8-20), ... | | Authors: | Ringel, A.E, Roman, C, Wolberger, C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Alternate deacylating specificities of the archaeal sirtuins Sir2Af1 and Sir2Af2.
Protein Sci., 23, 2014
|
|
4TWJ
 
 | | The structure of Sir2Af2 bound to a myristoylated histone peptide | | Descriptor: | ACETATE ION, GLYCEROL, Histone H4 peptide, ... | | Authors: | Ringel, A.E, Roman, C, Wolberger, C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Alternate deacylating specificities of the archaeal sirtuins Sir2Af1 and Sir2Af2.
Protein Sci., 23, 2014
|
|
1S7G
 
 | | Structural Basis for the Mechanism and Regulation of Sir2 Enzymes | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, HEXAETHYLENE GLYCOL, ... | | Authors: | Avalos, J.L, Boeke, J.D, Wolberger, C. | | Deposit date: | 2004-01-29 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the mechanism and regulation of sir2 enzymes
Mol.Cell, 13, 2004
|
|
6VEN
 
 | |
1AKH
 
 | | MAT A1/ALPHA2/DNA TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*AP*TP*GP*TP*AP*AP*AP*AP*AP*TP*TP*TP*AP*C P*AP*TP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*AP*TP*GP*TP*AP*AP*AP*TP*TP*TP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MATING-TYPE PROTEIN A-1), ... | | Authors: | Li, T, Jin, Y, Vershon, A.K, Wolberger, C. | | Deposit date: | 1997-05-19 | | Release date: | 1998-05-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the MATa1/MATalpha2 homeodomain heterodimer in complex with DNA containing an A-tract.
Nucleic Acids Res., 26, 1998
|
|
6AQR
 
 | | SAGA DUB module Ubp8(C146A)/Sgf11/Sus1/Sgf73 bound to monoubiquitin | | Descriptor: | Polyubiquitin-C, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Morrow, M.E, Morgan, M.T, Wolberger, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active site alanine mutations convert deubiquitinases into high-affinity ubiquitin-binding proteins.
EMBO Rep., 19, 2018
|
|
4ZUX
 
 | |
4LDT
 
 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBCH5B~Ub | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ubiquitin, ... | | Authors: | Wiener, R, DiBello, A.T, Lombardi, P.M, Guzzo, C.M, Zhang, X, Matunis, M.J, Wolberger, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
