1ZAR
 
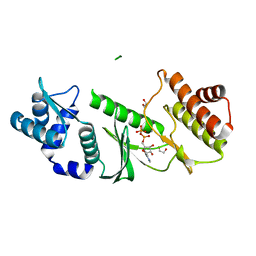 | | Crystal Structure of A.fulgidus Rio2 Kinase Complexed With ADP and Manganese Ions | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Laronde-Leblanc, N, Guszczynski, T, Copeland, T, Wlodawer, A. | | Deposit date: | 2005-04-06 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes.
Febs J., 272, 2005
|
|
1ZP8
 
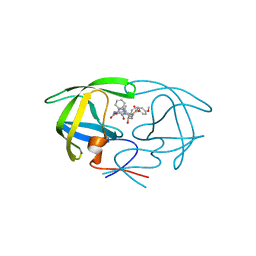 | | HIV Protease with inhibitor AB-2 | | Descriptor: | Pol polyprotein, [1-((1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL)-1H-1,2,3-TRIAZOL-4-YL]METHYL (1R,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YLCARBAMATE | | Authors: | Brik, A, Alexandratos, J.N, Elder, J.H, Olson, A.J, Wlodawer, A, Goodsell, D.S, Wong, C.H. | | Deposit date: | 2005-05-16 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | 1,2,3-triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors.
Chembiochem, 6, 2005
|
|
1ODY
 
 | | HIV-1 PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, HIV-1 PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
1L5E
 
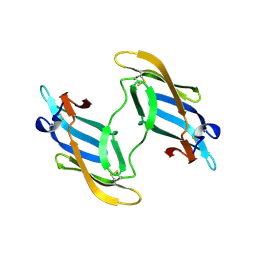 | | The domain-swapped dimer of CV-N in solution | | Descriptor: | Cyanovirin-N | | Authors: | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | Deposit date: | 2002-03-06 | | Release date: | 2002-06-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
1HFW
 
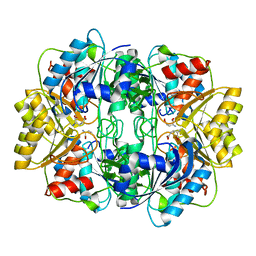 | |
1HFK
 
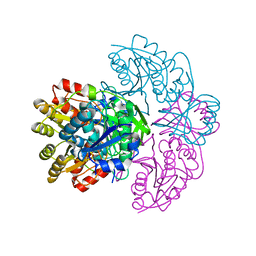 | | Asparaginase from Erwinia chrysanthemi, hexagonal form with weak sulfate | | Descriptor: | L-ASPARAGINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Lubkowski, J, Palm, G.J, Kozak, M, Jaskolski, M, Wlodawer, A. | | Deposit date: | 2000-12-05 | | Release date: | 2000-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structures of Two Highly Homologous Bacterial L-Asparaginases: A Case of Enantiomorphic Space Groups
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1YG9
 
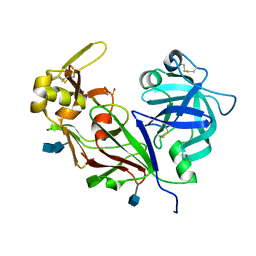 | | The structure of mutant (N93Q) of bla g 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aspartic protease Bla g 2, ... | | Authors: | Li, M, Gustchina, A, Wuenschmann, S, Pomes, A, Wlodawer, A. | | Deposit date: | 2005-01-04 | | Release date: | 2005-03-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of cockroach allergen Bla g 2, an unusual zinc binding aspartic protease with a novel mode of self-inhibition.
J.Mol.Biol., 348, 2005
|
|
1HFJ
 
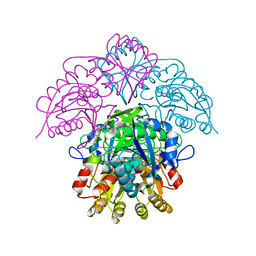 | | Asparaginase from Erwinia chrysanthemi, hexagonal form with sulfate | | Descriptor: | L-ASPARAGINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Palm, G.J, Lubkowski, J, Kozak, M, Jaskolski, M, Wlodawer, A. | | Deposit date: | 2000-12-05 | | Release date: | 2000-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Two Highly Homologous Bacterial L-Asparaginases: A Case of Enantiomorphic Space Groups
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HG1
 
 | |
1HG0
 
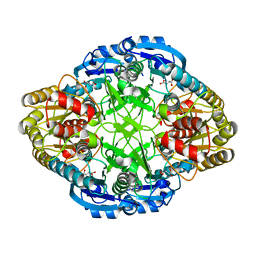 | |
1QDM
 
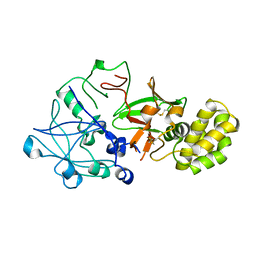 | | CRYSTAL STRUCTURE OF PROPHYTEPSIN, A ZYMOGEN OF A BARLEY VACUOLAR ASPARTIC PROTEINASE. | | Descriptor: | PROPHYTEPSIN | | Authors: | Kervinen, J, Tobin, G.J, Costa, J, Waugh, D.S, Wlodawer, A, Zdanov, A. | | Deposit date: | 1999-05-19 | | Release date: | 1999-07-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of plant aspartic proteinase prophytepsin: inactivation and vacuolar targeting.
EMBO J., 18, 1999
|
|
3LKY
 
 | | Monomeric Griffithsin with a Single Gly-Ser Insertion | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Griffithsin, ... | | Authors: | Moulaei, T, Wlodawer, A. | | Deposit date: | 2010-01-28 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity.
Structure, 18, 2010
|
|
3LL2
 
 | |
1ASW
 
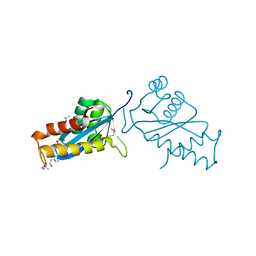 | | AVIAN SARCOMA VIRUS INTEGRASE CATALYTIC CORE DOMAIN CRYSTALLIZED FROM 20% PEG 4000, 10% ISOPROPANOL, HEPES PH 7.5 USING SELENOMETHIONINE SUBSTITUTED PROTEIN; DATA COLLECTED AT-165 DEGREES C | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE, ISOPROPYL ALCOHOL | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-08-25 | | Release date: | 1995-11-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structure of the catalytic domain of avian sarcoma virus integrase.
J.Mol.Biol., 253, 1995
|
|
1ASV
 
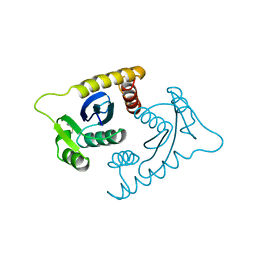 | |
1B3A
 
 | | TOTAL CHEMICAL SYNTHESIS AND HIGH-RESOLUTION CRYSTAL STRUCTURE OF THE POTENT ANTI-HIV PROTEIN AOP-RANTES | | Descriptor: | PENTYLOXYAMINO-ACETALDEHYDE, PROTEIN (RANTES), SULFATE ION | | Authors: | Wilken, J, Hoover, D, Thompson, D.A, Barlow, P.N, Mcsparron, H, Picard, L, Wlodawer, A, Lubkowski, J, Kent, S.B.H. | | Deposit date: | 1998-12-07 | | Release date: | 1999-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Total chemical synthesis and high-resolution crystal structure of the potent anti-HIV protein AOP-RANTES.
Chem.Biol., 6, 1999
|
|
1ASU
 
 | | AVIAN SARCOMA VIRUS INTEGRASE CATALYTIC CORE DOMAIN CRYSTALLIZED FROM 2% PEG 400, 2M AMMONIUM SULFATE, HEPES PH 7.5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-08-25 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structure of the catalytic domain of avian sarcoma virus integrase.
J.Mol.Biol., 253, 1995
|
|
1ZAO
 
 | | Crystal Structure of A.fulgidus Rio2 Kinase Complexed With ATP and Manganese Ions | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Laronde-Leblanc, N, Guszczynski, T, Copeland, T, Wlodawer, A. | | Deposit date: | 2005-04-06 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes.
Febs J., 272, 2005
|
|
1VSM
 
 | |
1VSE
 
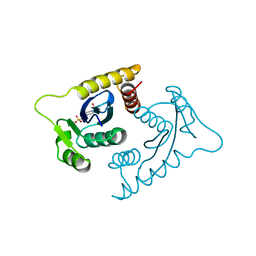 | | ASV INTEGRASE CORE DOMAIN WITH MG(II) COFACTOR AND HEPES LIGAND, LOW MG CONCENTRATION FORM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, INTEGRASE | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations.
Structure, 4, 1996
|
|
1VSF
 
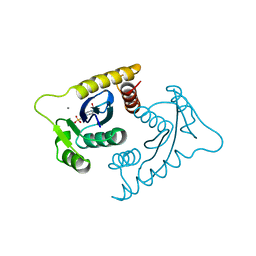 | | ASV INTEGRASE CORE DOMAIN WITH MN(II) COFACTOR AND HEPES LIGAND, HIGH MG CONCENTRATION FORM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, INTEGRASE, MANGANESE (II) ION | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations.
Structure, 4, 1996
|
|
1VSK
 
 | |
1PV8
 
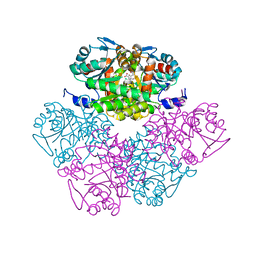 | | Crystal structure of a low activity F12L mutant of human porphobilinogen synthase | | Descriptor: | 3-(2-AMINOETHYL)-4-(AMINOMETHYL)HEPTANEDIOIC ACID, Delta-aminolevulinic acid dehydratase, ZINC ION | | Authors: | Breinig, S, Kervinen, J, Stith, L, Wasson, A.S, Fairman, R, Wlodawer, A, Zdanov, A, Jaffe, E.K. | | Deposit date: | 2003-06-26 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase.
Nat.Struct.Biol., 10, 2003
|
|
1TD0
 
 | | Viral capsid protein SHP at pH 5.5 | | Descriptor: | Head decoration protein | | Authors: | Chang, C, Forrer, P, Ott, D, Wlodawer, A, Plueckthun, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP
J.Mol.Biol., 344, 2004
|
|
2ANE
 
 | | Crystal structure of N-terminal domain of E.Coli Lon Protease | | Descriptor: | ATP-dependent protease La | | Authors: | Li, M, Rasulova, F, Melnikov, E.E, Rotanova, T.V, Gustchina, A, Maurizi, M.R, Wlodawer, A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the N-terminal domain of E. coli Lon protease.
Protein Sci., 14, 2005
|
|
