3C1Y
 
 | | Structure of bacterial DNA damage sensor protein with co-purified and co-crystallized ligand | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, DNA integrity scanning protein disA | | Authors: | Witte, G, Hartung, S, Buttner, K, Hopfner, K.P. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Biochemistry of a Bacterial Checkpoint Protein Reveals Diadenylate Cyclase Activity Regulated by DNA Recombination Intermediates
Mol.Cell, 30, 2008
|
|
3C21
 
 | | Structure of a bacterial DNA damage sensor protein with reaction product | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, DNA integrity scanning protein disA | | Authors: | Witte, G, Hartung, S, Buttner, K, Hopfner, K.P. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Biochemistry of a Bacterial Checkpoint Protein Reveals Diadenylate Cyclase Activity Regulated by DNA Recombination Intermediates
Mol.Cell, 30, 2008
|
|
3C23
 
 | | Structure of a bacterial DNA damage sensor protein with non-reactive Ligand | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA integrity scanning protein disA | | Authors: | Witte, G, Hartung, S, Buttner, K, Hopfner, K.P. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Biochemistry of a Bacterial Checkpoint Protein Reveals Diadenylate Cyclase Activity Regulated by DNA Recombination Intermediates
Mol.Cell, 30, 2008
|
|
3C1Z
 
 | |
4NQI
 
 | | Structure of the N-terminal I-BAR domain (1-259) of D.Discoideum IBARa | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, SH3 domain-containing protein | | Authors: | Witte, G, Faix, J, Runge-Wollmann, P. | | Deposit date: | 2013-11-25 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The inverse BAR domain protein IBARa drives membrane remodeling to control osmoregulation, phagocytosis and cytokinesis.
J.Cell.Sci., 127, 2014
|
|
5O58
 
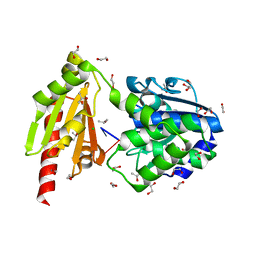 | | Structure of the inactive T.maritima PDE (TM1595) D80N D154N mutant with substrate 5'-pApG | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-06-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
5O4Z
 
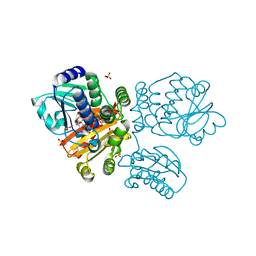 | | Structure of the inactive T.maritima PDE (TM1595) D80N D154N mutant with substrate 5'-pApA | | Descriptor: | ADENOSINE-5'-MONOPHOSPHATE, CHLORIDE ION, DHH/DHHA1-type phosphodiesterase TM1595, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-05-31 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
5O7F
 
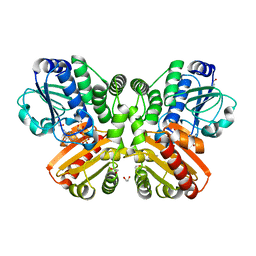 | | Structure of the inactive T.maritima PDE (TM1595) D80N D154N mutant with GMP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
5O1U
 
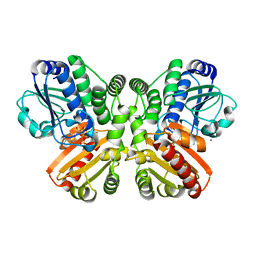 | | Structure of wildtype T.maritima PDE (TM1595) with AMP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-05-19 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
5O70
 
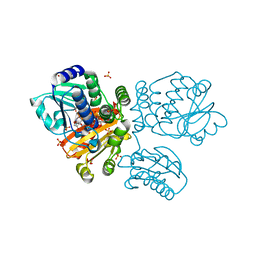 | |
5O25
 
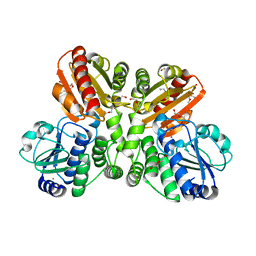 | | Structure of wildtype T.maritima PDE (TM1595) in ligand-free state | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-05-19 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
7A08
 
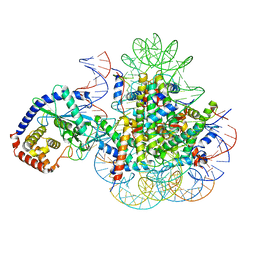 | | CryoEM Structure of cGAS Nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, Histone H2A type 1-C, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Michalski, S, de Oliveira Mann, C.C, Witte, G, Bartho, J, Lammens, K, Hopfner, K.P. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for sequestration and autoinhibition of cGAS by chromatin.
Nature, 587, 2020
|
|
7OZ3
 
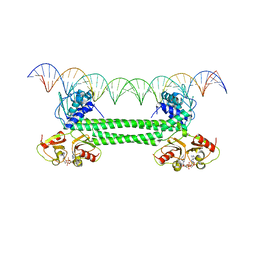 | | S. agalactiae BusR in complex with its busA-promotor DNA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GntR family transcriptional regulator, pBusA_for, ... | | Authors: | Bandera, A.M, Witte, G. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res., 49, 2021
|
|
4WK1
 
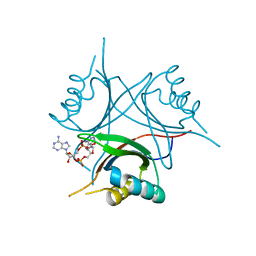 | | Crystal structure of Staphylococcus aureus PstA in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CALCIUM ION, PstA | | Authors: | Mueller, M, Hopfner, K.-P, Witte, G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | c-di-AMP recognition by Staphylococcus aureus PstA.
Febs Lett., 589, 2015
|
|
4WK3
 
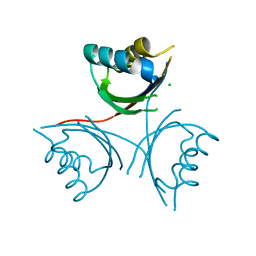 | |
5EOM
 
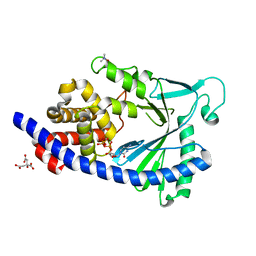 | | Structure of full-length human MAB21L1 with bound CTP | | Descriptor: | CITRIC ACID, CYTIDINE-5'-TRIPHOSPHATE, Protein mab-21-like 1, ... | | Authors: | de Oliveira Mann, C.C, Witte, G, Hopfner, K.-P. | | Deposit date: | 2015-11-10 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and biochemical characterization of the cell fate determining nucleotidyltransferase fold protein MAB21L1.
Sci Rep, 6, 2016
|
|
5EOG
 
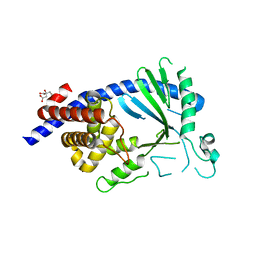 | |
3OC3
 
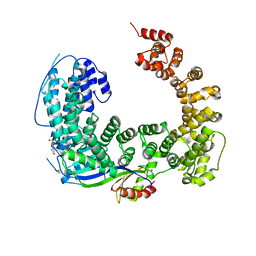 | | Crystal structure of the Mot1 N-terminal domain in complex with TBP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HELICASE MOT1, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Wollmann, P, Cui, S, Viswanathan, R, Berninghausen, O, Wells, M.N, Moldt, M, Witte, G, Butryn, A, Wendler, P, Beckmann, R, Auble, D.T, Hopfner, K.-P. | | Deposit date: | 2010-08-09 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
4YVZ
 
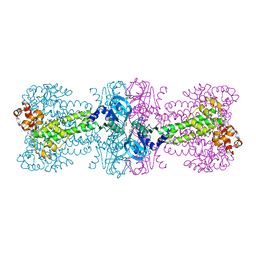 | | Structure of Thermotoga maritima DisA in complex with 3'-dATP/Mn2+ | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA integrity scanning protein DisA, MANGANESE (II) ION | | Authors: | Mueller, M, Deimling, T, Hopfner, K.-P, Witte, G. | | Deposit date: | 2015-03-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Structural analysis of the diadenylate cyclase reaction of DNA-integrity scanning protein A (DisA) and its inhibition by 3'-dATP.
Biochem.J., 469, 2015
|
|
3TJ1
 
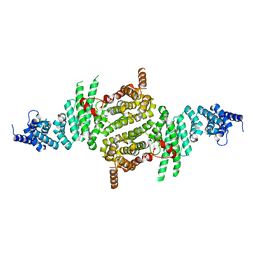 | | Crystal Structure of RNA Polymerase I Transcription Initiation Factor Rrn3 | | Descriptor: | RNA polymerase I-specific transcription initiation factor RRN3 | | Authors: | Blattner, C, Jennebach, S, Herzog, F, Mayer, A, Cheung, A.C.M, Witte, G, Lorenzen, K, Hopfner, K.-P, Heck, A.J.R, Aebersold, R, Cramer, P. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth.
Genes Dev., 25, 2011
|
|
4YXJ
 
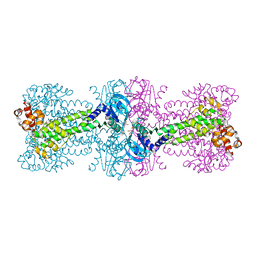 | | Structure of Thermotoga maritima DisA in complex with ApCpp | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA integrity scanning protein DisA | | Authors: | Mueller, M, Deimling, T, Hopfner, K.-P, Witte, G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural analysis of the diadenylate cyclase reaction of DNA-integrity scanning protein A (DisA) and its inhibition by 3'-dATP.
Biochem.J., 469, 2015
|
|
4YXM
 
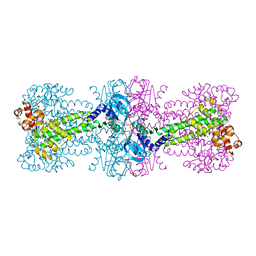 | | Structure of Thermotoga maritima DisA D75N mutant with reaction product c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA integrity scanning protein DisA | | Authors: | Mueller, M, Deimling, T, Hopfner, K.-P, Witte, G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of the diadenylate cyclase reaction of DNA-integrity scanning protein A (DisA) and its inhibition by 3'-dATP.
Biochem.J., 469, 2015
|
|
4ON9
 
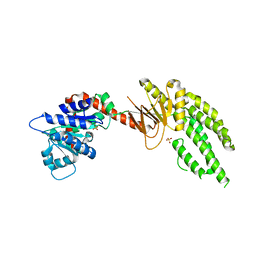 | | DECH box helicase domain | | Descriptor: | CHLORIDE ION, Probable ATP-dependent RNA helicase DDX58, SULFATE ION | | Authors: | Deimling, T, Witte, G, Hopfner, K.P. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal and solution structure of the human RIG-I SF2 domain
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3QB0
 
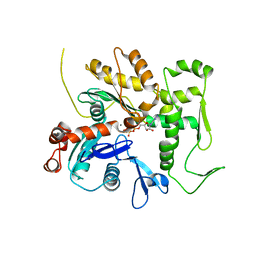 | | Crystal structure of Actin-related protein Arp4 from S. cerevisiae complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 4, CALCIUM ION | | Authors: | Fenn, S, Breitsprecher, D, Gerhold, C.B, Witte, G, Faix, J, Hopfner, K.P. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Structural biochemistry of nuclear actin-related proteins 4 and 8 reveals their interaction with actin.
Embo J., 30, 2011
|
|
3PP5
 
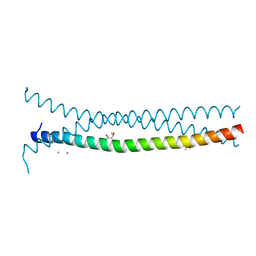 | | High-resolution structure of the trimeric Scar/WAVE complex precursor Brk1 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Brk1, ... | | Authors: | Linkner, J, Witte, G, Curth, U, Faix, J. | | Deposit date: | 2010-11-24 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution X-ray structure of the trimeric Scar/WAVE-complex precursor Brk1.
Plos One, 6, 2011
|
|
