2NEF
 
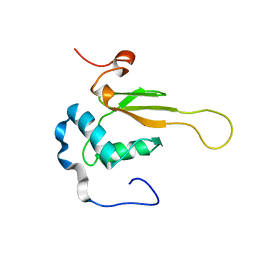 | | HIV-1 NEF (REGULATORY FACTOR), NMR, 40 STRUCTURES | | Descriptor: | NEGATIVE FACTOR (F-PROTEIN) | | Authors: | Grzesiek, S, Bax, A, Clore, G.M, Gronenborn, A.M, Hu, J.S, Kaufman, J, Palmer, I, Stahl, S.J, Tjandra, N, Wingfield, P.T. | | Deposit date: | 1997-02-12 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure and backbone dynamics of HIV-1 Nef.
Protein Sci., 6, 1997
|
|
6UI7
 
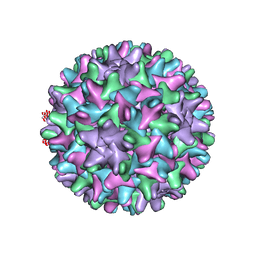 | | HBV T=4 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
2X7L
 
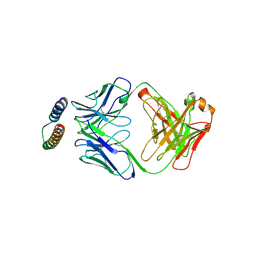 | | Implications of the HIV-1 Rev dimer structure at 3.2A resolution for multimeric binding to the Rev response element | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, PROTEIN REV | | Authors: | DiMattia, M.A, Watts, N.R, Stahl, S.J, Rader, C, Wingfield, P.T, Stuart, D.I, Steven, A.C, Grimes, J.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Implications of the HIV-1 Rev Dimer Structure at 3. 2 A Resolution for Multimeric Binding to the Rev Response Element.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6UI6
 
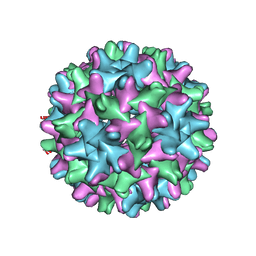 | | HBV T=3 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
2HGF
 
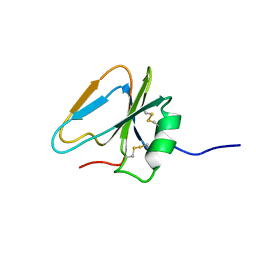 | | HAIRPIN LOOP CONTAINING DOMAIN OF HEPATOCYTE GROWTH FACTOR, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEPATOCYTE GROWTH FACTOR | | Authors: | Zhou, H, Mazzulla, M.J, Kaufman, J.D, Stahl, S.J, Wingfield, P.T, Rubin, J.S, Bottaro, D.P, Byrd, R.A. | | Deposit date: | 1997-12-18 | | Release date: | 1998-06-24 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of hepatocyte growth factor reveals a potential heparin-binding site.
Structure, 6, 1998
|
|
4RVJ
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with amprenavir | | Descriptor: | HIV-1 protease, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2014-11-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced antiviral activity by the P2-tris-tetrahydrofuran moiety of GRL-0519, a novel nonpeptidic HIV-1 protease inhibitor (PI), against multi-PI-resistant and highly darunavir-resistant strains of HIV-1
To be Published
|
|
4RVI
 
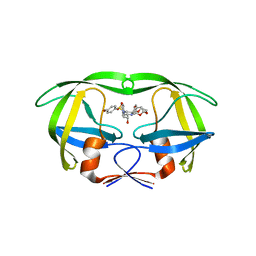 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2014-11-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Enhanced antiviral activity by the P2-tris-tetrahydrofuran moiety of GRL-0519, a novel nonpeptidic HIV-1 protease inhibitor (PI), against multi-PI-resistant and highly darunavir-resistant strains of HIV-1
To be Published
|
|
4RVX
 
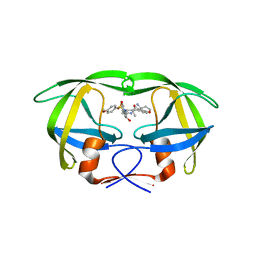 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL079 | | Descriptor: | (4S)-4-amino-N-[(2S,3S)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]-3,4-dihydro-2H-chromene-6-carboxamide, HIV-1 Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2014-11-28 | | Release date: | 2016-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Enhanced antiviral activity by the P2-tris-tetrahydrofuran moiety of GRL-0519, a novel nonpeptidic HIV-1 protease inhibitor (PI), against multi-PI-resistant and highly darunavir-resistant strains of HIV-1.
To be Published
|
|
4HLA
 
 | | Crystal structure of wild type HIV-1 protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2012-10-16 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
4I8W
 
 | | Crystal structure of wild type HIV-1 protease in complex with non-peptidic inhibitor, GRL007 | | Descriptor: | 4-{[(2R,3S)-3-({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yloxy]carbonyl}amino)-2-hydroxy-4-phenylbutyl](2-methylpropyl)sulfamoyl}benzoic acid, Protease | | Authors: | Yedidi, R.S, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2012-12-04 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
4I8Z
 
 | | Crystal structure of wild type HIV-1 protease in complex with non-peptidic inhibitor, GRL008 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-carbamoylphenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2012-12-04 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
3V6Z
 
 | | Crystal Structure of Hepatitis B Virus e-antigen | | Descriptor: | Fab e6 Heavy Chain, Fab e6 Light Chain, e-antigen | | Authors: | Dimattia, M.A, Watts, N.R, Stahl, S.J, Grimes, J.M, Steven, A.C, Stuart, D.I, Wingfield, P.T. | | Deposit date: | 2011-12-20 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein.
Structure, 21, 2013
|
|
3V6F
 
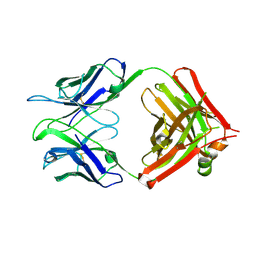 | | Crystal Structure of an anti-HBV e-antigen monoclonal Fab fragment (e6), unbound | | Descriptor: | Fab e6 Heavy Chain, Fab e6 Light Chain | | Authors: | Dimattia, M.A, Watts, N.R, Stahl, S.J, Grimes, J.M, Steven, A.C, Stuart, D.I, Wingfield, P.T. | | Deposit date: | 2011-12-19 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein.
Structure, 21, 2013
|
|
2ARI
 
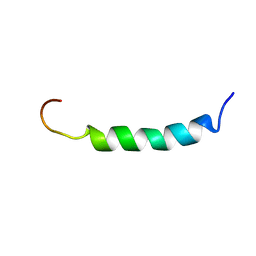 | | Solution structure of micelle-bound fusion domain of HIV-1 gp41 | | Descriptor: | Envelope polyprotein GP160 | | Authors: | Jaroniec, C.P, Kaufman, J.D, Stahl, S.J, Viard, M, Blumenthal, R, Wingfield, P.T, Bax, A. | | Deposit date: | 2005-08-19 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Micelle-Associated Human Immunodeficiency Virus gp41 Fusion Domain.
Biochemistry, 44, 2005
|
|
1QBZ
 
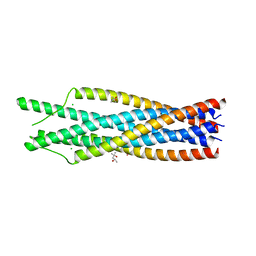 | | THE CRYSTAL STRUCTURE OF THE SIV GP41 ECTODOMAIN AT 1.47 A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, MERCURY (II) ION, ... | | Authors: | Yang, Z.-N, Mueser, T.C, Kaufman, J, Stahl, S.J, Wingfield, P.T, Hyde, C.C. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The crystal structure of the SIV gp41 ectodomain at 1.47 A resolution.
J.Struct.Biol., 126, 1999
|
|
5DHV
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
5DHZ
 
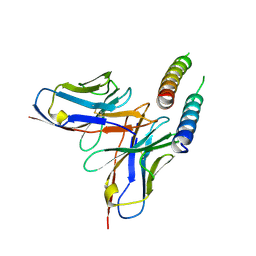 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
5DHX
 
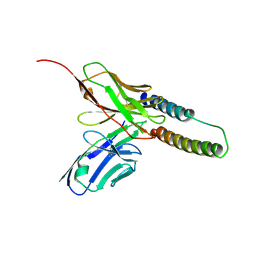 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, light chain,Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
5DHY
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
1EY1
 
 | | SOLUTION STRUCTURE OF ESCHERICHIA COLI NUSB | | Descriptor: | ANTITERMINATION FACTOR NUSB | | Authors: | Altieri, A.S, Mazzulla, M.J, Horita, D.A, Coats, R.H, Wingfield, P.T, Byrd, R.A. | | Deposit date: | 2000-05-05 | | Release date: | 2000-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the transcriptional antiterminator NusB from Escherichia coli.
Nat.Struct.Biol., 7, 2000
|
|
6BSY
 
 | | HIV-1 Rev assembly domain (residues 1-69) | | Descriptor: | PHOSPHATE ION, Protein Rev | | Authors: | Watts, N.R, Eren, E, Zhuang, X, Wang, Y.X, Steven, A.C, Wingfield, P.T. | | Deposit date: | 2017-12-04 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A new HIV-1 Rev structure optimizes interaction with target RNA (RRE) for nuclear export.
J. Struct. Biol., 203, 2018
|
|
6CF2
 
 | | Crystal structure of HIV-1 Rev (residues 1-93)-RNA aptamer complex | | Descriptor: | Anti-Rev Antibody, heavy chain, light chain, ... | | Authors: | Eren, E, Dearborn, A.D, Wingfield, P.T. | | Deposit date: | 2018-02-13 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an RNA Aptamer that Can Inhibit HIV-1 by Blocking Rev-Cognate RNA (RRE) Binding and Rev-Rev Association.
Structure, 26, 2018
|
|
4NJU
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with tipranavir | | Descriptor: | N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NJV
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with ritonavir | | Descriptor: | Protease, RITONAVIR | | Authors: | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
6CWT
 
 | |
