2O4Z
 
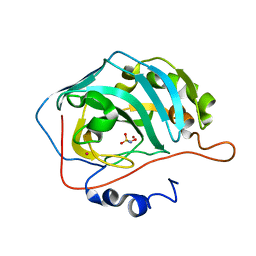 | | Crystal structure of the Carbonic Anhydrase II complexed with hydroxysulfamide inhibitor | | Descriptor: | Carbonic anhydrase 2, MERCURY (II) ION, N-hydroxysulfamide, ... | | Authors: | Temperini, C, Winum, J.Y, Montero, J.L, Scozzafava, a, Supuran, c.t. | | Deposit date: | 2006-12-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbonic anhydrase inhibitors: The X-ray crystal structure of the adduct of N-hydroxysulfamide with isozyme II explains why this new zinc binding function is effective in the design of potent inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3MJD
 
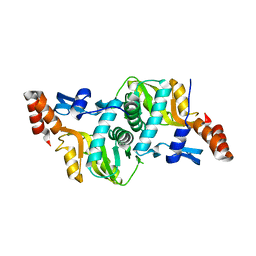 | | 1.9 Angstrom Crystal Structure of Orotate Phosphoribosyltransferase (pyrE) Francisella tularensis. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Orotate phosphoribosyltransferase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-12 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Orotate Phosphoribosyltransferase (pyrE) Francisella tularensis.
TO BE PUBLISHED
|
|
4KWT
 
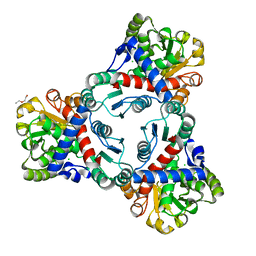 | | Crystal structure of unliganded anabolic ornithine carbamoyltransferase from Vibrio vulnificus at 1.86 A resolution | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Ornithine carbamoyltransferase | | Authors: | Shabalin, I.G, Bacal, P, Bajor, J, Winsor, J, Grimshaw, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-05-24 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
To be Published
|
|
3RJ5
 
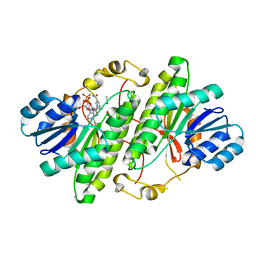 | | Structure of alcohol dehydrogenase from Drosophila lebanonesis T114V mutant complexed with NAD+ | | Descriptor: | ACETIC ACID, Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Morgunova, E, Wuxiuer, Y, Cols, N, Popov, A, Sylte, I, Karshikoff, A, Gonzales-Duarte, R, Ladenstein, R, Winberg, J.O. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An intact eight-membered water chain in drosophilid alcohol dehydrogenases is essential for optimal enzyme activity.
Febs J., 279, 2012
|
|
4MVA
 
 | | 1.43 Angstrom Resolution Crystal Structure of Triosephosphate Isomerase (tpiA) from Escherichia coli in Complex with Acetyl Phosphate. | | Descriptor: | 1,2-ETHANEDIOL, ACETYLPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Kuhn, M.L, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
Plos One, 9, 2014
|
|
4JFR
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with carbamoyl phosphate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Ornithine carbamoyltransferase, ... | | Authors: | Shabalin, I.G, Winsor, J, Grimshaw, S, Osinski, T, Bajor, J, Chordia, M.D, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
to be published
|
|
1PIY
 
 | | RIBONUCLEOTIDE REDUCTASE R2 SOAKED WITH FERROUS ION AT NEUTRAL PH | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
1PIM
 
 | | DITHIONITE REDUCED E. COLI RIBONUCLEOTIDE REDUCTASE R2 SUBUNIT, D84E MUTANT | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Khidekel, N, Baldwin, J, Ley, B.A, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Ribonucleotide Reductase R2 Mutant that Accumulates a u-1,2-Peroxodiiron(III)
Intermediate during Oxygen Activation
J.Am.Chem.Soc., 122, 2000
|
|
3N2B
 
 | | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae. | | Descriptor: | CHLORIDE ION, Diaminopimelate decarboxylase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae.
To be Published
|
|
1PJ0
 
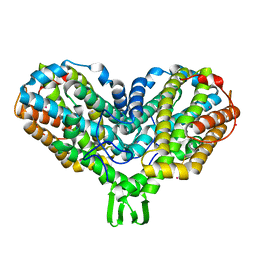 | | RIBONUCLEOTIDE REDUCTASE R2-D84E/W48F MUTANT SOAKED WITH FERROUS IONS AT NEUTRAL PH | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
1PIZ
 
 | | RIBONUCLEOTIDE REDUCTASE R2 D84E MUTANT SOAKED WITH FERROUS IONS AT NEUTRAL PH | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
1PIU
 
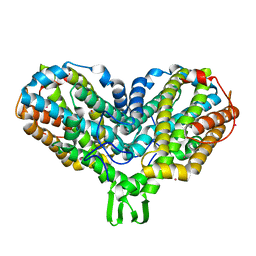 | | OXIDIZED RIBONUCLEOTIDE REDUCTASE R2-D84E MUTANT CONTAINING OXO-BRIDGED DIFERRIC CLUSTER | | Descriptor: | FE (III) ION, MERCURY (II) ION, OXYGEN ATOM, ... | | Authors: | Voegtli, W.C, Khidekel, N, Baldwin, J, Ley, B.A, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Ribonucleotide Reductase R2 Mutant that Accumulates a u-1,2-Peroxodiiron(III)
Intermediate during Oxygen Activation
J.Am.Chem.Soc., 122, 2000
|
|
1PJ1
 
 | | RIBONUCLEOTIDE REDUCTASE R2-D84E/W48F SOAKED WITH FERROUS IONS AT PH 5 | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
1Q40
 
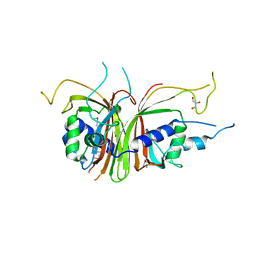 | | Crystal structure of the C. albicans Mtr2-Mex67 M domain complex | | Descriptor: | GLYCEROL, MRNA TRANSPORT REGULATOR Mtr2, mRNA export factor MEX67 | | Authors: | Senay, C, Ferrari, P, Rocher, C, Rieger, K.J, Winter, J, Platel, D, Bourne, Y. | | Deposit date: | 2003-08-01 | | Release date: | 2003-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The mtr2-mex67 ntf2-like domain complex: Structural
insights into a dual role of MTR2 for yeast nuclear export
J.Biol.Chem., 278, 2003
|
|
1Q42
 
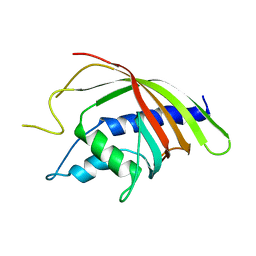 | | Crystal structure analysis of the Candida albicans Mtr2 | | Descriptor: | MRNA TRANSPORT REGULATOR Mtr2 | | Authors: | Senay, C, Ferrari, P, Rocher, C, Rieger, K.J, Winter, J, Platel, D, Bourne, Y. | | Deposit date: | 2003-08-01 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The mtr2-mex67 ntf2-like domain complex: Structural
insights into a dual role of MTR2 for yeast nuclear export
J.Biol.Chem., 278, 2003
|
|
3NZT
 
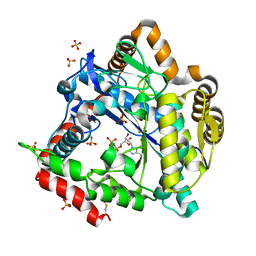 | | 2.0 Angstrom Crystal structure of Glutamate--Cysteine Ligase (gshA) ftom Francisella tularensis in Complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glutamate--cysteine ligase, SULFATE ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-16 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Crystal structure of Glutamate--Cysteine Ligase (gshA) ftom Francisella tularensis in Complex with AMP.
To be Published
|
|
3O2R
 
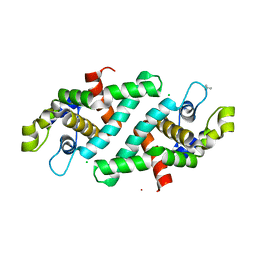 | | Structural flexibility in region involved in dimer formation of nuclease domain of Ribonuclase III (rnc) from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, Ribonuclease III | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structural Flexibility in Region Involved in Dimer Formation of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni.
TO BE PUBLISHED
|
|
1PM2
 
 | | CRYSTAL STRUCTURE OF MANGANESE SUBSTITUTED R2-D84E (D84E MUTANT OF THE R2 SUBUNIT OF E. COLI RIBONUCLEOTIDE REDUCTASE) | | Descriptor: | MANGANESE (II) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Baldwin, J, Saleh, L, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-06-09 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
4NOI
 
 | | 2.17 Angstrom Crystal Structure of DNA-directed RNA Polymerase Subunit Alpha from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, DNA-directed RNA polymerase subunit alpha, IODIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | 2.17 Angstrom Crystal Structure of DNA-directed RNA Polymerase Subunit Alpha from Campylobacter jejuni.
TO BE PUBLISHED
|
|
4NV4
 
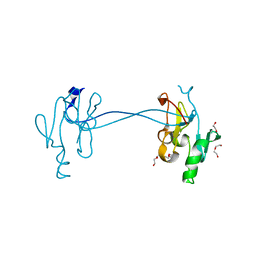 | | 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Signal peptidase I, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis.
TO BE PUBLISHED
|
|
4GIB
 
 | | 2.27 Angstrom Crystal Structure of beta-Phosphoglucomutase (pgmB) from Clostridium difficile | | Descriptor: | Beta-phosphoglucomutase, GLYCINE, PHOSPHATE ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | 2.27 Angstrom Crystal Structure of beta-Phosphoglucomutase (pgmB) from Clostridium difficile.
TO BE PUBLISHED
|
|
3HJB
 
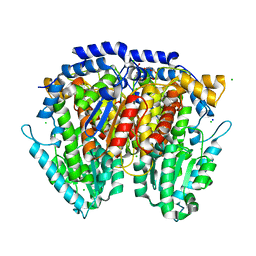 | | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae.
To be Published
|
|
4DBU
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 (AKR1C3) in complex with NADP+ and 3-((4 -(trifluoromethyl)phenyl)amino)benzoic acid | | Descriptor: | 3-{[4-(trifluoromethyl)phenyl]amino}benzoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Chen, M, Christianson, D.W, Winkler, J.D, Penning, T.M. | | Deposit date: | 2012-01-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.528 Å) | | Cite: | Crystal structures of AKR1C3 containing an N-(aryl)amino-benzoate inhibitor and a bifunctional AKR1C3 inhibitor and androgen receptor antagonist. Therapeutic leads for castrate resistant prostate cancer.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3HVU
 
 | | 1.95 Angstrom Crystal Structure of Complex of Hypoxanthine-Guanine Phosphoribosyltransferase from Bacillus anthracis with 2-(N-morpholino)ethanesulfonic acid (MES) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hypoxanthine phosphoribosyltransferase, SODIUM ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-16 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Complex of Hypoxanthine-Guanine Phosphoribosyltransferase from Bacillus anthracis with 2-(N-morpholino)ethanesulfonic acid (MES)
TO BE PUBLISHED
|
|
4O4A
 
 | | 2.75 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis. | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, Lipoprotein, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Winsor, J, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis.
TO BE PUBLISHED
|
|
