1A48
 
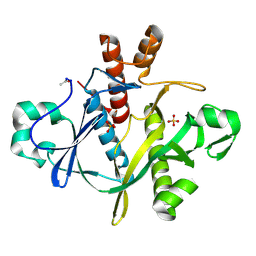 | | SAICAR SYNTHASE | | Descriptor: | PHOSPHORIBOSYLAMINOIMIDAZOLE-SUCCINOCARBOXAMIDE SYNTHASE, SULFATE ION | | Authors: | Levdikov, V.M, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1998-02-12 | | Release date: | 1999-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of SAICAR synthase: an enzyme in the de novo pathway of purine nucleotide biosynthesis.
Structure, 6, 1998
|
|
1AYX
 
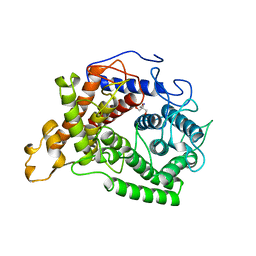 | | CRYSTAL STRUCTURE OF GLUCOAMYLASE FROM SACCHAROMYCOPSIS FIBULIGERA AT 1.7 ANGSTROMS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUCOAMYLASE | | Authors: | Sevcik, J, Hostinova, E, Gasperik, J, Solovicova, A, Wilson, K.S, Dauter, Z. | | Deposit date: | 1997-11-12 | | Release date: | 1998-05-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of glucoamylase from Saccharomycopsis fibuligera at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AY7
 
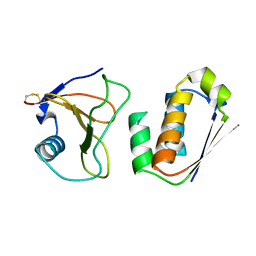 | | RIBONUCLEASE SA COMPLEX WITH BARSTAR | | Descriptor: | BARSTAR, GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Sevcik, J, Urbanikova, L, Dauter, Z, Wilson, K.S. | | Deposit date: | 1997-11-14 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of RNase Sa by the inhibitor barstar: structure of the complex at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1BLU
 
 | | STRUCTURE OF THE 2[4FE-4S] FERREDOXIN FROM CHROMATIUM VINOSUM | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Moulis, J.M. | | Deposit date: | 1996-04-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the 2[4Fe-4S] ferredoxin from Chromatium vinosum: evolutionary and mechanistic inferences for [3/4Fe-4S] ferredoxins.
Protein Sci., 5, 1996
|
|
1BOX
 
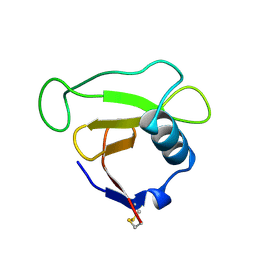 | | N39S MUTANT OF RNASE SA FROM STREPTOMYCES AUREOFACIENS | | Descriptor: | GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Hebert, E.J, Giletto, A, Sevcik, J, Urbanikova, L, Wilson, K.S, Dauter, Z, Pace, C.N. | | Deposit date: | 1998-08-07 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Contribution of a conserved asparagine to the conformational stability of ribonucleases Sa, Ba, and T1.
Biochemistry, 37, 1998
|
|
1E3Z
 
 | | Acarbose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.93A | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1DUC
 
 | | EIAV DUTPASE DUDP/STRONTIUM COMPLEX | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, DEOXYURIDINE-5'-DIPHOSPHATE, STRONTIUM ION | | Authors: | Dauter, Z, Persson, R, Rosengren, A.M, Nyman, P.O, Wilson, K.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 1997-11-29 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex.
J.Mol.Biol., 285, 1999
|
|
1E43
 
 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.7A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1DUN
 
 | | EIAV DUTPASE NATIVE | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Dauter, Z, Persson, R, Rosengren, A.M, Nyman, P.O, Wilson, K.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 1997-11-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex.
J.Mol.Biol., 285, 1999
|
|
1E40
 
 | | Tris/maltotriose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 2.2A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1E3X
 
 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.92A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-26 | | Release date: | 2001-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1EB6
 
 | | Deuterolysin from Aspergillus oryzae | | Descriptor: | 1,2-ETHANEDIOL, NEUTRAL PROTEASE II, ZINC ION | | Authors: | McAuley, K.E, Jia-Xing, Y, Dodson, E.J, Lehmbeck, J, Ostergaard, P.R, Wilson, K.S. | | Deposit date: | 2001-07-19 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Quick Solution: Ab Initio Structure Determination of a 19 kDa Metalloproteinase Using Acorn
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1PEK
 
 | | STRUCTURE OF THE COMPLEX OF PROTEINASE K WITH A SUBSTRATE-ANALOGUE HEXA-PEPTIDE INHIBITOR AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | D-DAL-ALA-NH2, PEPTIDE PRO-ALA-PRO-PHE, PROTEINASE K | | Authors: | Betzel, C, Singh, T.P, Visanji, M, Peters, K, Fittkau, S, Saenger, W, Wilson, K.S. | | Deposit date: | 1993-01-19 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of proteinase K with a substrate analogue hexapeptide inhibitor at 2.2-A resolution.
J.Biol.Chem., 268, 1993
|
|
1PJ8
 
 | | Structure of a ternary complex of proteinase K, mercury and a substrate-analogue hexapeptide at 2.2 A resolution | | Descriptor: | 6-residue peptide (N-Ac-PAPFPA-NH2), MERCURY (II) ION, Proteinase K | | Authors: | Saxena, A.K, Singh, T.P, Peters, K, Fittkau, S, Visanji, M, Wilson, K.S, Betzel, C. | | Deposit date: | 2003-06-02 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a ternary complex of proteinase K, mercury, and a substrate-analogue hexa-peptide at 2.2 A resolution
Proteins, 25, 1996
|
|
1QBA
 
 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1QH7
 
 | | CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE | | Descriptor: | XYLANASE, beta-D-xylopyranose | | Authors: | Sabini, E, Sulzenbacher, G, Dauter, M, Dauter, Z, Jorgensen, P.L, Schulein, M, Dupont, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase.
Chem.Biol., 6, 1999
|
|
1QHP
 
 | | FIVE-DOMAIN ALPHA-AMYLASE FROM BACILLUS STEAROTHERMOPHILUS, MALTOSE COMPLEX | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Dauter, Z, Dauter, M, Brzozowski, A.M, Christensen, S, Borchert, T.V, Beier, L, Wilson, K.S, Davies, G.J. | | Deposit date: | 1999-05-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of Novamyl, the five-domain "maltogenic" alpha-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7A resolution.
Biochemistry, 38, 1999
|
|
1QNR
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, GLYCEROL, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNQ
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2,2':6',2''-TERPYRIDINE PLATINUM(II) Chloride, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1OTG
 
 | | 5-CARBOXYMETHYL-2-HYDROXYMUCONATE ISOMERASE | | Descriptor: | 5-CARBOXYMETHYL-2-HYDROXYMUCONATE ISOMERASE, SULFATE ION | | Authors: | Subramanya, H.S, Roper, D.I, Dauter, Z, Dodson, E.J, Davies, G.J, Wilson, K.S, Wigley, D.B. | | Deposit date: | 1995-11-09 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases.
Biochemistry, 35, 1996
|
|
1OGL
 
 | | The crystal structure of native Trypanosoma cruzi dUTPase | | Descriptor: | DEOXYURIDINE TRIPHOSPHATASE | | Authors: | Harkiolaki, M, Dodson, E.J, Bernier-Villamor, V, Turkenburg, J.P, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2003-05-07 | | Release date: | 2004-01-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Dutpase Reveals a Novel Dutp/Dudp Binding Fold
Structure, 12, 2004
|
|
1OGK
 
 | | The crystal structure of Trypanosoma cruzi dUTPase in complex with dUDP | | Descriptor: | DEOXYURIDINE TRIPHOSPHATASE, DEOXYURIDINE-5'-DIPHOSPHATE | | Authors: | Harkiolaki, M, Dodson, E.J, Bernier-Villamor, V, Turkenburg, J.P, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2003-05-07 | | Release date: | 2004-01-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Dutpase Reveals a Novel Dutp/Dudp Binding Fold
Structure, 12, 2004
|
|
1QBB
 
 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-07 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1OPO
 
 | | THE STRUCTURE OF CARNATION MOTTLE VIRUS | | Descriptor: | CALCIUM ION, Coat protein, SULFATE ION | | Authors: | Morgunova, E, Dauter, Z, Fry, E, Stuart, D, Stel'mashchuk, V, Mikhailov, A.M, Wilson, K.S, Vainshtein, B.K. | | Deposit date: | 2003-03-06 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The atomic structure of Carnation Mottle Virus capsid protein
Febs Lett., 338, 1994
|
|
1QNO
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
