2V1R
 
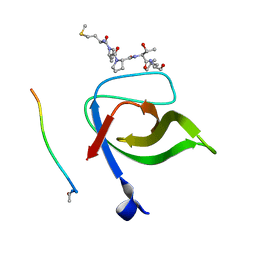 | | Yeast Pex13 SH3 domain complexed with a peptide from Pex14 at 2.1 A resolution | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PAS20, PEX14 | | Authors: | Kursula, I, Kursula, P, Lehmann, F, Zou, P, Song, Y.H, Wilmanns, M. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Genomics of Yeast SH3 Domains
To be Published
|
|
2VKN
 
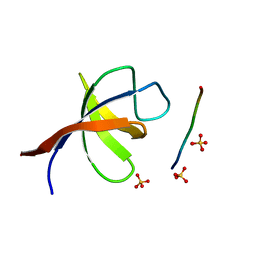 | | YEAST SHO1 SH3 DOMAIN COMPLEXED WITH A PEPTIDE FROM PBS2 | | Descriptor: | MAP KINASE KINASE PBS2, PROTEIN SSU81, SULFATE ION | | Authors: | Kursula, P, Kursula, I, Song, Y.H, Paraskevopoulos, K, Wilmanns, M. | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Genomics of Yeast SH3 Domains
To be Published
|
|
2V1Q
 
 | | Atomic-resolution structure of the yeast Sla1 SH3 domain 3 | | Descriptor: | CHLORIDE ION, CYTOSKELETON ASSEMBLY CONTROL PROTEIN SLA1, PLATINUM (II) ION, ... | | Authors: | Kursula, I, Kursula, P, Zou, P, Lehmann, F, Song, Y.H, Wilmanns, M. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Genomics of Yeast SH3 Domains
To be Published
|
|
2W4J
 
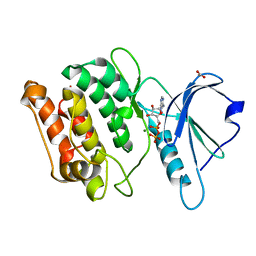 | | X-ray structure of a DAP-Kinase 2-277 | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, DEATH-ASSOCIATED PROTEIN KINASE 1, ... | | Authors: | De Diego, I, Kuper, J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2008-11-27 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-Ray Structure of a Dap-Kinase Calmodulin Complex
To be Published
|
|
2W4K
 
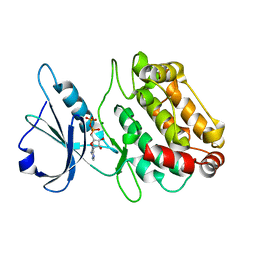 | | X-ray structure of a DAP-Kinase 2-302 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DEATH-ASSOCIATED PROTEIN KINASE 1, MAGNESIUM ION | | Authors: | De Diego, I, Kuper, J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2008-11-27 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Pef/Y Substrate Recognition and Signature Motif Plays a Critical Role in Dapk-Related Kinase Activity.
Chem.Biol., 21, 2014
|
|
2WT7
 
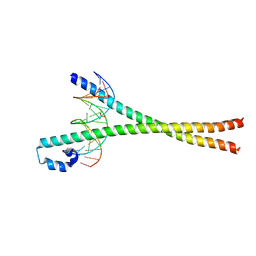 | | Crystal structure of the bZIP heterodimeric complex MafB:cFos bound to DNA | | Descriptor: | MODIFIED T-MARE MOTIF, PHOSPHATE ION, PROTO-ONCOGENE PROTEIN C-FOS, ... | | Authors: | Pogenberg, V, Holton, S, Wilmanns, M. | | Deposit date: | 2009-09-11 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of a bZIP Transcription Factor with Homo/Heterodimer-Induced DNA-Binding Preference.
Structure, 22, 2014
|
|
2WJZ
 
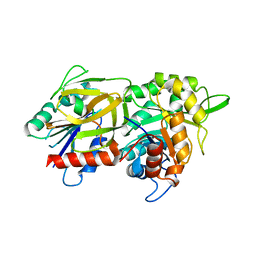 | | Crystal structure of (HisH) K181A Y138A mutant of imidazoleglycerolphosphate synthase (HisH HisF) which displays constitutive glutaminase activity | | Descriptor: | IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE HISF, IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISH, PHOSPHATE ION | | Authors: | Vega, M.C, List, F, Razeto, A, Haeger, M.C, Babinger, K, Kuper, J, Sterner, R, Wilmanns, M. | | Deposit date: | 2009-06-02 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Catalysis Uncoupling in a Glutamine Amidotransferase Bienzyme by Unblocking the Glutaminase Active Site.
Chem.Biol., 19, 2012
|
|
8ARI
 
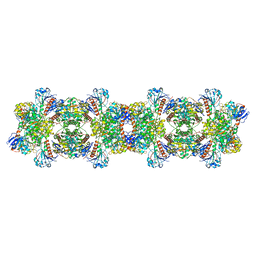 | |
8ATI
 
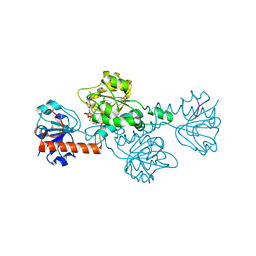 | |
8BX1
 
 | |
8BX2
 
 | |
1UUE
 
 | | a-SPECTRIN SH3 DOMAIN (V44T, D48G MUTANT) | | Descriptor: | SPECTRIN ALPHA CHAIN | | Authors: | Vega, M.C, Fernandez, A, Wilmanns, M, Serrano, L. | | Deposit date: | 2003-12-18 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Solvation in Protein Folding Analysis: Combination of Theoretical and Experimental Approaches
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1THF
 
 | |
1UU0
 
 | | Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (Apo-form) | | Descriptor: | HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Vega, M.C, Fernandez, F.J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
1UU2
 
 | | Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (apo-form) | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, HISTIDINOL-PHOSPHATE AMINOTRANSFERASE | | Authors: | Vega, M.C, Fernandez, F.J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2003-12-13 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
1UU1
 
 | | Complex of Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (Apo-form) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PHOSPHORIC ACID MONO-[2-AMINO-3-(3H-IMIDAZOL-4-YL)-PROPYL]ESTER | | Authors: | Vega, M.C, Fernandez, F.J, Lehman, F, Wilmanns, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
2Y9M
 
 | | Pex4p-Pex22p structure | | Descriptor: | 1,2-ETHANEDIOL, PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-10-26 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into Ubiquitin-Conjugating Enzyme/ Co-Activator Interactions from the Structure of the Pex4P:Pex22P Complex.
Embo J., 31, 2011
|
|
2YQ4
 
 | |
1TG0
 
 | |
1SSH
 
 | | Crystal structure of the SH3 domain from a S. cerevisiae hypothetical 40.4 kDa protein in complex with a peptide | | Descriptor: | 12-mer peptide from Cytoskeleton assembly control protein SLA1, Hypothetical 40.4 kDa protein in PES4-HIS2 intergenic region | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Song, Y.-H, Wilmanns, M. | | Deposit date: | 2004-03-24 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Yeast SH3 domain structural genomics
To be Published
|
|
1SYL
 
 | | Crystal structure of inactive mutant dUTPase complexed with substrate dUTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Kovari, J, Pongracz, V, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2004-04-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase
J.Biol.Chem., 279, 2004
|
|
2Y9P
 
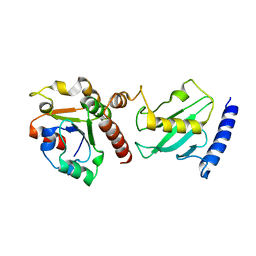 | | Pex4p-Pex22p mutant II structure | | Descriptor: | PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Pex4P-Pex22P Structure
To be Published
|
|
1RUW
 
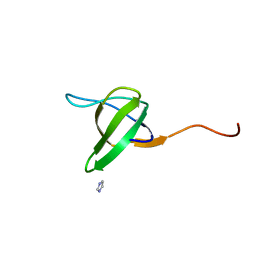 | | Crystal structure of the SH3 domain from S. cerevisiae Myo3 | | Descriptor: | IMIDAZOLE, Myosin-3 isoform | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Song, Y.H, Wilmanns, M. | | Deposit date: | 2003-12-12 | | Release date: | 2005-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the SH3 domain from S. cerevisiae Myo3
To be Published
|
|
2YQ5
 
 | | Crystal Structure of D-isomer specific 2-hydroxyacid dehydrogenase from Lactobacillus delbrueckii ssp. bulgaricus: NAD complexed form | | Descriptor: | D-ISOMER SPECIFIC 2-HYDROXYACID DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Holton, S.J, Anandhakrishnan, M, Geerlof, A, Wilmanns, M. | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Characterization of D-Isomer Specific 2-Hydroxyacid Dehydrogenase from Lactobacillus Delbrueckii Ssp. Bulgaricus
J.Struct.Biol., 181, 2013
|
|
1RN8
 
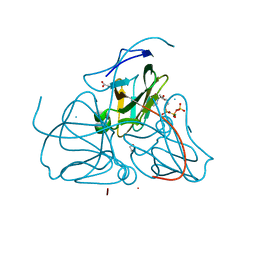 | | Crystal structure of dUTPase complexed with substrate analogue imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
