1W2F
 
 | | Human Inositol (1,4,5)-trisphosphate 3-kinase substituted with selenomethionine | | Descriptor: | INOSITOL-TRISPHOSPHATE 3-KINASE A, SULFATE ION | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
8AJ8
 
 | | Structure of p110 gamma bound to the p84 regulatory subunit | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, Phosphoinositide 3-kinase regulatory subunit 6 | | Authors: | Burke, J.E, Williams, R.L, Zhang, X. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (8.5 Å) | | Cite: | Molecular basis for differential activation of p101 and p84 complexes of PI3K gamma by Ras and GPCRs.
Cell Rep, 42, 2023
|
|
2XVC
 
 | | Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division | | Descriptor: | CADMIUM ION, CDVA, SSO0911, ... | | Authors: | Samson, R.Y, Obita, T, Hodgson, B, Shaw, M.K, Chong, P.L, Williams, R.L, Bell, S.D. | | Deposit date: | 2010-10-25 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular and Structural Basis of Escrt-III Recruitment to Membranes During Archaeal Cell Division.
Mol.Cell, 41, 2011
|
|
5FVM
 
 | | Cryo electron microscopy of a complex of Tor and Lst8 | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE TOR2, TARGET OF RAPAMYCIN COMPLEX SUBUNIT LST8 | | Authors: | Baretic, D, Berndt, A, Ohashi, Y, Johnson, C.M, Williams, R.L. | | Deposit date: | 2016-02-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Tor Forms a Dimer Through an N-Terminal Helical Solenoid with a Complex Topology
Nat.Commun., 7, 2016
|
|
5WPU
 
 | | Crystal structure HpiC1 Y101S | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WPS
 
 | | Crystal structure HpiC1 Y101F | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5KC1
 
 | | Structure of the C-terminal dimerization domain of Atg38 | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Autophagy-related protein 38, ... | | Authors: | Ohashi, Y, Soler, N, Garcia-Ortegon, M, Zhang, L, Perisic, O, Masson, G.R, Johnson, C.M, Williams, R.J. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
2PT7
 
 | | Crystal structure of Cag VirB11 (HP0525) and an inhibitory protein (HP1451) | | Descriptor: | Cag-alfa, Hypothetical protein | | Authors: | Hare, S, Fischer, W, Williams, R, Terradot, L, Bayliss, R, Haas, R, Waksman, G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification, structure and mode of action of a new regulator of the Helicobacter pylori HP0525 ATPase.
Embo J., 26, 2007
|
|
5WPR
 
 | | Crystal structure HpiC1 in C2 space group | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WPP
 
 | | Crystal structure HpiC1 W73M/K132M | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
2CAY
 
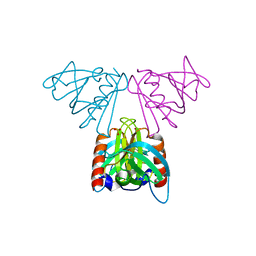 | | Vps36 N-terminal PH domain | | Descriptor: | SULFATE ION, VACUOLAR PROTEIN SORTING PROTEIN 36 | | Authors: | Teo, H, Williams, R.L, Perisic, O, Gill, D.J. | | Deposit date: | 2005-12-23 | | Release date: | 2006-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Escrt-I Core and Escrt-II Glue Domain Structures Reveal Role for Glue in Linking to Escrt-I and Membranes.
Cell(Cambridge,Mass.), 125, 2006
|
|
2CAZ
 
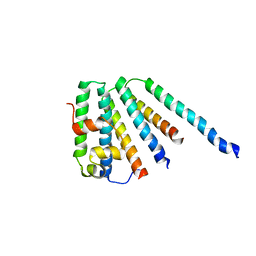 | | ESCRT-I core | | Descriptor: | PROTEIN SRN2, SUPPRESSOR PROTEIN STP22 OF TEMPERATURE-SENSITIVE ALPHA-FACTOR RECEPTOR AND ARGININE PERMEASE, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN VPS28 | | Authors: | Gill, D.J, Teo, H, Sun, J, Perisic, O, Veprintsev, D.B, Vallis, Y, Emr, S.D, Williams, R.L. | | Deposit date: | 2005-12-23 | | Release date: | 2006-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Escrt-I Core and Escrt-II Glue Domain Structures Reveal Role for Glue in Linking to Escrt-I and Membranes.
Cell(Cambridge,Mass.), 125, 2006
|
|
1TLG
 
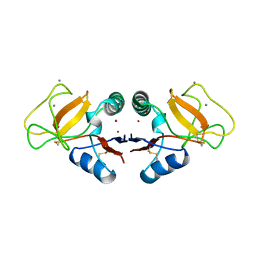 | | STRUCTURE OF A TUNICATE C-TYPE LECTIN COMPLEXED WITH D-GALACTOSE | | Descriptor: | CALCIUM ION, POLYANDROCARPA LECTIN, ZINC ION, ... | | Authors: | Poget, S.F, Legge, G.B, Bycroft, M, Williams, R.L. | | Deposit date: | 1998-10-16 | | Release date: | 1999-07-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a tunicate C-type lectin from Polyandrocarpa misakiensis complexed with D -galactose.
J.Mol.Biol., 290, 1999
|
|
2R1Z
 
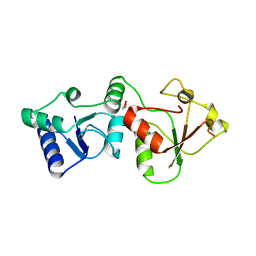 | |
2G50
 
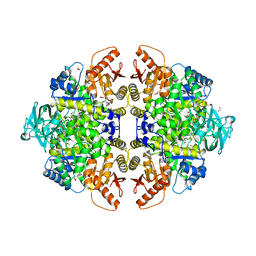 | | The location of the allosteric amino acid binding site of muscle pyruvate kinase. | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ALANINE, ... | | Authors: | Holyoak, T, Williams, R, Fenton, A.W. | | Deposit date: | 2006-02-22 | | Release date: | 2006-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differentiating a Ligand's Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase.
Biochemistry, 45, 2006
|
|
2DOQ
 
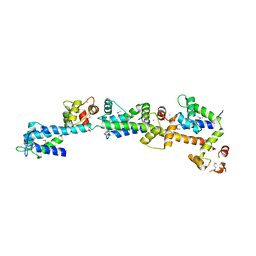 | | crystal structure of Sfi1p/Cdc31p complex | | Descriptor: | CALCIUM ION, Cell division control protein 31, SFI1p | | Authors: | Li, S, Sandercock, A.M, Conduit, P.T, Robinson, C.V, Williams, R.L, Kilmartin, J.V. | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication.
J.Cell Biol., 173, 2006
|
|
2GV5
 
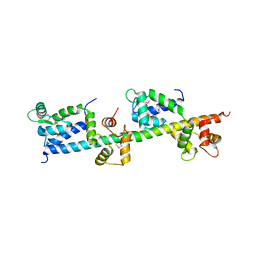 | | crystal structure of Sfi1p/Cdc31p complex | | Descriptor: | Cell division control protein 31, Sfi1p | | Authors: | Li, S, Sandercock, A.M, Conduit, P.T, Robinson, C.V, Williams, R.L, Kilmartin, J.V. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication.
J.Cell Biol., 173, 2006
|
|
6Q00
 
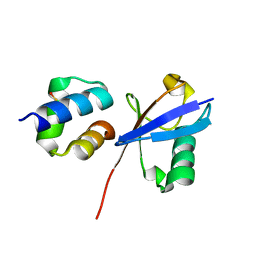 | | TDP2 UBA Domain Bound to Ubiquitin at 0.85 Angstroms Resolution, Crystal Form 1 | | Descriptor: | POTASSIUM ION, Tyrosyl-DNA phosphodiesterase 2, Ubiquitin | | Authors: | Schellenberg, M.J, Krahn, J.M, Williams, R.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.
Nucleic Acids Res., 48, 2020
|
|
1UZX
 
 | | A complex of the Vps23 UEV with ubiquitin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UBIQUITIN, ... | | Authors: | Teo, H, Williams, R.L. | | Deposit date: | 2004-03-18 | | Release date: | 2004-03-30 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Endosomal Sorting Complex Required for Transport (Escrt-I) Recognition of Ubiquitinated Proteins
J.Biol.Chem., 279, 2004
|
|
2YMB
 
 | | Structures of MITD1 | | Descriptor: | CHARGED MULTIVESICULAR BODY PROTEIN 1A, MIT DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Hadders, M.A, Agromayor, M, Obita, T, Perisic, O, Caballe, A, Kloc, M, Lamers, M.H, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1YJ5
 
 | | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme | | Descriptor: | 5' polynucleotide kinase-3' phosphatase FHA domain, 5' polynucleotide kinase-3' phosphatase catalytic domain, SULFATE ION | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-13 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
1YJM
 
 | | Crystal structure of the FHA domain of mouse polynucleotide kinase in complex with an XRCC4-derived phosphopeptide. | | Descriptor: | 12-mer peptide from DNA-repair protein XRCC4, Polynucleotide 5'-hydroxyl-kinase | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
4GZ0
 
 | |
8BFU
 
 | | Crystal structure of the apo p110alpha catalytic subunit from homo sapiens | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gong, G.Q, Bellini, D, Vanhaesebroeck, B, Williams, R.L. | | Deposit date: | 2022-10-26 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
4GYZ
 
 | | Mus Musculus Tdp2 Bound to dAMP and Mg2+ | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Williams, R.S. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.556 Å) | | Cite: | Mechanism of repair of 5'-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
