6TG5
 
 | |
1R9K
 
 | | Representative solution structure of the catalytic domain of SopE2 | | Descriptor: | TypeIII-secreted protein effector: invasion-associated protein | | Authors: | Williams, C, Galyov, E.E, Bagby, S. | | Deposit date: | 2003-10-30 | | Release date: | 2004-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Backbone Dynamics, and Interaction with Cdc42 of Salmonella Guanine Nucleotide Exchange Factor SopE2(,).
Biochemistry, 43, 2004
|
|
1R6E
 
 | |
1L5J
 
 | | CRYSTAL STRUCTURE OF E. COLI ACONITASE B. | | Descriptor: | ACONITATE ION, Aconitate hydratase 2, FE3-S4 CLUSTER | | Authors: | Williams, C.H, Stillman, T.J, Barynin, V.V, Sedelnikova, S.E, Tang, Y, Green, J, Guest, J.R, Artymiuk, P.J. | | Deposit date: | 2002-03-07 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | E. coli aconitase B structure reveals a HEAT-like domain with implications for protein-protein recognition.
Nat.Struct.Biol., 9, 2002
|
|
2L21
 
 | | chicken IGF2R domain 11 | | Descriptor: | Cation-independent mannose-6-phosphate receptor | | Authors: | Williams, C, Hoppe, H, Strickland, M, Frago, S, Ellis, R.Z, Wattana-Amorn, P, Prince, S.N, Zaccheo, O.J, Forbes, B, Jones, E.Y, Rezgui, D.Z, Crump, M.P, Hassan, A.B. | | Deposit date: | 2010-08-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | CD loop dependency of the IGF2: M6P/IGF2 receptor binding interaction predates imprinting
To be Published
|
|
2CNJ
 
 | | NMR studies on the interaction of Insulin-Growth Factor II (IGF-II) with IGF2R domain 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Williams, C, Prince, S, Zaccheo, O, Hassan, A.B, Crosby, J, Crump, M. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights Into the Interaction of Insulin-Like Growth Factor 2 with Igf2R Domain 11.
Structure, 15, 2007
|
|
2Y9P
 
 | | Pex4p-Pex22p mutant II structure | | Descriptor: | PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Pex4P-Pex22P Structure
To be Published
|
|
2Y9M
 
 | | Pex4p-Pex22p structure | | Descriptor: | 1,2-ETHANEDIOL, PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-10-26 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into Ubiquitin-Conjugating Enzyme/ Co-Activator Interactions from the Structure of the Pex4P:Pex22P Complex.
Embo J., 31, 2011
|
|
2L29
 
 | | Complex structure of E4 mutant human IGF2R domain 11 bound to IGF-II | | Descriptor: | Insulin-like growth factor 2 receptor variant, Insulin-like growth factor II | | Authors: | Williams, C, Hoppe, H, Rezgui, D, Strickland, M, Frago, S, Ellis, R.Z, Wattana-Amorn, P, Prince, S.N, Zaccheo, O.J, Forbes, B, Jones, E.Y, Crump, M.P, Hassan, A.B. | | Deposit date: | 2010-08-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | An exon splice enhancer primes IGF2:IGF2R binding site structure and function evolution.
Science, 338, 2012
|
|
2L2A
 
 | | Mutated Domain 11 of the Cytoplasmic region of the Cation-independent mannose-6-phosphate receptor | | Descriptor: | Insulin-like growth factor 2 receptor variant | | Authors: | Williams, C, Hoppe, H, Rezgui, D, Strickland, M, Frago, S, Ellis, R.Z, Wattana-Amorn, P, Prince, S.N, Zaccheo, O.J, Forbes, B, Jones, E.Y, Crump, M.P, Hassan, A.B. | | Deposit date: | 2010-08-13 | | Release date: | 2012-02-15 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | An exon splice enhancer primes IGF2:IGF2R binding site structure and function evolution.
Science, 338, 2012
|
|
2L2G
 
 | | Solution structure of Opossum Domain 11 | | Descriptor: | IGF2R DOMAIN 11 | | Authors: | Williams, C, Hoppe, H, Rezgui, D, Rezgui, M, Frago, S, Ellis, R.Z, Wattana-Amorn, P, Prince, S.N, Zaccheo, O.J, Forbes, B, Jones, E.Y, Crump, M.P, Bassim, A.H. | | Deposit date: | 2010-08-18 | | Release date: | 2012-02-15 | | Last modified: | 2015-02-25 | | Method: | SOLUTION NMR | | Cite: | An exon splice enhancer primes IGF2:IGF2R binding site structure and function evolution.
Science, 338, 2012
|
|
4BWF
 
 | | Pex4p-Pex22p disulphide bond mutant | | Descriptor: | 1,2-ETHANEDIOL, PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Stanley, W.A, Wilmanns, M, Distel, B. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | A Disulphide Bond in the E2 Enzyme Pex4P Modulates Ubiquitin-Conjugating Activity
Sci.Rep., 3, 2013
|
|
2PNG
 
 | | Type I rat fatty acid synthase acyl carrier protein (ACP) domain | | Descriptor: | Fatty acid synthase (EC 2.3.1.85) | | Authors: | Ploskon, E.A, Arthur, C.J, Evans, S.E, Williams, C, Crosby, J, Crump, M.P. | | Deposit date: | 2007-04-24 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Mammalian type I Fatty Acid synthase acyl carrier protein domain does not sequester acyl chains.
J.Biol.Chem., 283, 2008
|
|
5WIG
 
 | |
8OL8
 
 | |
8OJR
 
 | | Solution NMR Structure of Alpha-Synuclein 1-25 Peptide in 50% TFE. | | Descriptor: | Alpha-synuclein | | Authors: | Allen, S.G, Williams, C, Meade, R.M, Tang, T.M.S, Crump, M.P, Mason, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-08-09 | | Method: | SOLUTION NMR | | Cite: | An N-terminal alpha-Synuclein fragment binds lipid vesicles to modulate lipid induced aggregation
Cell Rep Phys Sci, 2023
|
|
5LO4
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPa-CH3 | | Authors: | Baker, E.G, Williams, C, Hudson, K.L, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
6Z32
 
 | | Human cation-independent mannose 6-phosphate/IGF2 receptor domains 7-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, SULFATE ION, ... | | Authors: | Bochel, A.J, Williams, C, McCoy, A.J, Hoppe, H, Winter, A.J, Nicholls, R.D, Harlos, K, Jones, Y.E, Berger, I, Hassan, B, Crump, M.P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
6Z30
 
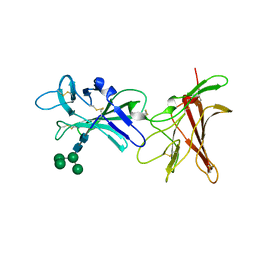 | | Human cation-independent mannose 6-phosphate/ IGF2 receptor domains 9-10 | | Descriptor: | Cation-independent mannose-6-phosphate receptor, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bochel, A.J, Williams, C, Crump, M.P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
6Z31
 
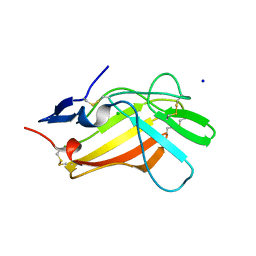 | |
6GWX
 
 | | Stabilising and Understanding a Miniprotein by Rational Design. | | Descriptor: | Optimised PPa-TYR | | Authors: | Porter Goff, K.L, Williams, C, Baker, E.G, Nicol, D, Samphire, J.L, Zieleniewski, F.L, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2018-06-26 | | Release date: | 2019-07-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Stabilizing and Understanding a Miniprotein by Rational Redesign.
Biochemistry, 58, 2019
|
|
1ATG
 
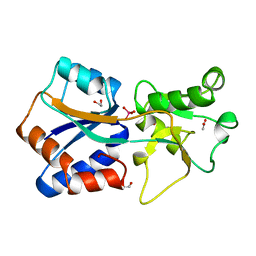 | | AZOTOBACTER VINELANDII PERIPLASMIC MOLYBDATE-BINDING PROTEIN | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PERIPLASMIC MOLYBDATE-BINDING PROTEIN, ... | | Authors: | Lawson, D.M, Pau, R.N, Williams, C.E.M, Mitchenall, L.A. | | Deposit date: | 1997-08-14 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 A resolution crystal structure of Azotobacter vinelandii ModA.
Structure, 6, 1998
|
|
5NKZ
 
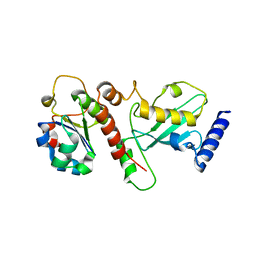 | | Crystal structure of H. polymorpha ubiquitin conjugating enzyme Pex4p in complex with soluble domain of Pex22p | | Descriptor: | Peroxin 22, Ubiquitin-conjugating enzyme E2-21 kDa | | Authors: | Danda, N, Lunev, S, Ali, A, Groves, M.R, Williams, C. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into K48-linked ubiquitin chain formation by the Pex4p-Pex22p complex.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
7OUB
 
 | |
5LO2
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPaTyr | | Authors: | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
