1VPU
 
 | |
1WBR
 
 | |
3ZUA
 
 | | A C39-like domain | | Descriptor: | ALPHA-HEMOLYSIN TRANSLOCATION ATP-BINDING PROTEIN HLYB | | Authors: | Lecher, J, Schwarz, C.K.W, Stoldt, M, Smits, S.S.H, Willbold, D, Schmitt, L. | | Deposit date: | 2011-07-18 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An Rtx Transporter Tethers its Unfolded Substrate During Secretion Via a Unique N-Terminal Domain.
Structure, 20, 2012
|
|
5O6F
 
 | | NMR structure of cold shock protein A from Corynebacterium pseudotuberculosis | | Descriptor: | Cold-shock protein | | Authors: | Caruso, I.P, Panwalkar, V, Coronado, M.A, Dingley, A.J, Cornelio, M.L, Willbold, D, Arni, R.K, Eberle, R.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-07-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and interaction of Corynebacterium pseudotuberculosis cold shock protein A with Y-box single-stranded DNA fragment.
FEBS J., 285, 2018
|
|
6Y1A
 
 | | Amyloid fibril structure of islet amyloid polypeptide | | Descriptor: | AMINO GROUP, Islet amyloid polypeptide | | Authors: | Roeder, C, Kupreichyk, T, Gremer, L, Schaefer, L.U, Pothula, K.R, Ravelli, R.B.G, Willbold, D, Hoyer, W, Schroder, G.F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-03-04 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of islet amyloid polypeptide fibrils reveals similarities with amyloid-beta fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6R4R
 
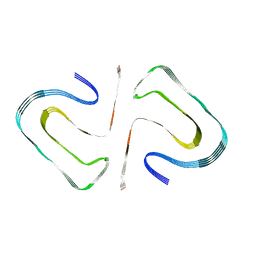 | | Cryo-EM Structure of the PI3-Kinase SH3 Domain Amyloid Fibril | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha | | Authors: | Roeder, C, Vettore, N, Mangels, L.N, Gremer, L, Ravelli, R.B.G, Willbold, D, Hoyer, W, Buell, A.K, Schroder, G.F. | | Deposit date: | 2019-03-23 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structure of PI3-kinase SH3 amyloid fibrils by cryo-electron microscopy.
Nat Commun, 10, 2019
|
|
1FI0
 
 | |
5OQV
 
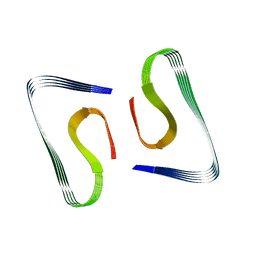 | | Near-atomic resolution fibril structure of complete amyloid-beta(1-42) by cryo-EM | | Descriptor: | Amyloid beta A4 protein | | Authors: | Gremer, L, Schoelzel, D, Schenk, C, Reinartz, E, Labahn, J, Ravelli, R, Tusche, M, Lopez-Iglesias, C, Hoyer, W, Heise, H, Willbold, D, Schroeder, G.F. | | Deposit date: | 2017-08-14 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Fibril structure of amyloid-beta (1-42) by cryo-electron microscopy.
Science, 358, 2017
|
|
8S1M
 
 | |
1TVT
 
 | |
4OV0
 
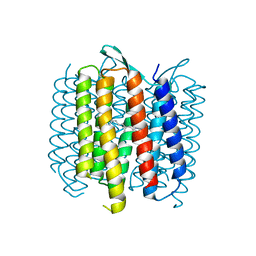 | | Structure of Bacteriorhdopsin Transferred from Amphipol A8-35 to a Lipidic Mesophase | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Polovinkin, V, Gushchin, I, Sintsov, M, Round, E, Balandin, T, Chervakov, P, Schevchenko, V, Utrobin, P, Popov, A, Borshchevskiy, V, Mishin, A, Kuklin, A, Willbold, D, Popot, J.L, Gordeliy, V. | | Deposit date: | 2014-02-19 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a membrane protein transferred from amphipol to a lipidic mesophase.
J.Membr.Biol., 247, 2014
|
|
2YOM
 
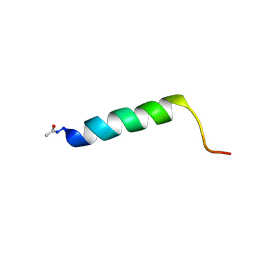 | | Solution NMR structure of the C-terminal extension of two bacterial light, oxygen, voltage (LOV) photoreceptor proteins from Pseudomonas putida | | Descriptor: | SENSORY BOX PROTEIN | | Authors: | Rani, R, Lecher, J, Hartmann, R, Krauss, U, Jaeger, K, Willbold, D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Conservation of Dark Recovery Kinetic Parameters and Structural Features in the Pseudomonadaceae "Short" Light, Oxygen, Voltage (Lov) Protein Family: Implications for the Design of Lov-Based Optogenetic Tools.
Biochemistry, 52, 2013
|
|
2YON
 
 | | Solution NMR structure of the C-terminal extension of two bacterial light, oxygen, voltage (LOV) photoreceptor proteins from Pseudomonas putida | | Descriptor: | SENSORY BOX PROTEIN | | Authors: | Rani, R, Hartmann, R, Lecher, J, Krauss, U, Jaeger, K, Willbold, D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Conservation of Dark Recovery Kinetic Parameters and Structural Features in the Pseudomonadaceae "Short" Light, Oxygen, Voltage (Lov) Protein Family: Implications for the Design of Lov-Based Optogenetic Tools.
Biochemistry, 52, 2013
|
|
2K0G
 
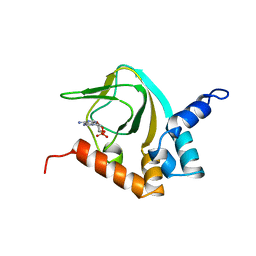 | |
2K7Y
 
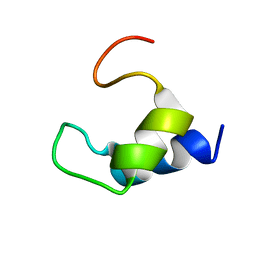 | |
4CO7
 
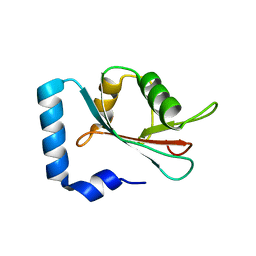 | | Crystal structure of human GATE-16 | | Descriptor: | GAMMA-AMINOBUTYRIC ACID RECEPTOR-ASSOCIATED PROTEIN-LIKE 2 | | Authors: | Weiergraeber, O.H, Ma, P, Willbold, D. | | Deposit date: | 2014-01-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Polymorphism in Autophagy-Related Protein Gate-16.
Biochemistry, 54, 2015
|
|
5K5G
 
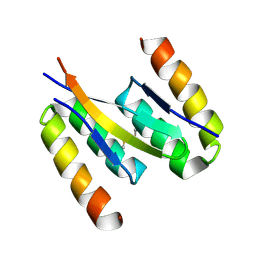 | | Structure of human islet amyloid polypeptide in complex with an engineered binding protein | | Descriptor: | HI18, Islet amyloid polypeptide | | Authors: | Mirecka, E.A, Feuerstein, S, Gremer, L, Schroeder, G.F, Stoldt, M, Willbold, D, Hoyer, W. | | Deposit date: | 2016-05-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | beta-Hairpin of Islet Amyloid Polypeptide Bound to an Aggregation Inhibitor.
Sci Rep, 6, 2016
|
|
1YO4
 
 | |
3D32
 
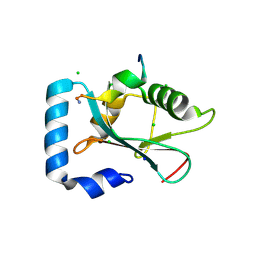 | | Complex of GABA(A) receptor-associated protein (GABARAP) with a synthetic peptide | | Descriptor: | CHLORIDE ION, Gamma-aminobutyric acid receptor-associated protein, K1 peptide, ... | | Authors: | Weiergraeber, O.H, Stangler, T, Willbold, D. | | Deposit date: | 2008-05-09 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Ligand Binding Mode of GABA(A) Receptor-Associated Protein.
J.Mol.Biol., 381, 2008
|
|
3DOW
 
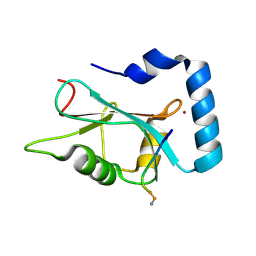 | |
4KUO
 
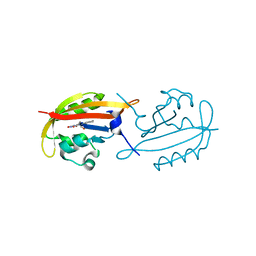 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Photoexcited state) | | Descriptor: | RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
3IQD
 
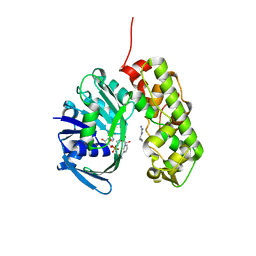 | | Structure of Octopine-dehydrogenase in complex with NADH and Agmatine | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, AGMATINE, Octopine dehydrogenase | | Authors: | Smits, S.H.J, Meyer, T, Mueller, A, Willbold, D, Grieshaber, M.K, Schmitt, L. | | Deposit date: | 2009-08-20 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into the mechanism of ligand binding to octopine dehydrogenase from Pecten maximus by NMR and crystallography
Plos One, 5, 2010
|
|
4KUK
 
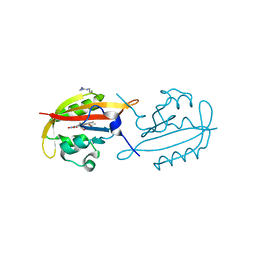 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Dark state) | | Descriptor: | ACETIC ACID, RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
1KIK
 
 | | SH3 Domain of Lymphocyte Specific Kinase (LCK) | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK | | Authors: | Briese, L, Willbold, D. | | Deposit date: | 2001-12-03 | | Release date: | 2001-12-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human Lck unique and SH3 domains by nuclear magnetic resonance spectroscopy.
Bmc Struct.Biol., 3, 2003
|
|
1KOT
 
 | |
