8BPT
 
 | |
5LOJ
 
 | |
5LOE
 
 | | Structure of full length Cody from Bacillus subtilis in complex with Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, ISOLEUCINE | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
5LNH
 
 | | Structure of full length Unliganded CodY from Bacillus subtilis | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, SULFATE ION | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
5LOO
 
 | |
2GX5
 
 | | N-terminal GAF domain of transcriptional pleiotropic repressor CodY | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, GTP-sensing transcriptional pleiotropic repressor codY, ... | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2006-05-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria.
J.Biol.Chem., 281, 2006
|
|
2HGV
 
 | |
1QKB
 
 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KVK | | Descriptor: | ACETATE ION, PEPTIDE LYS-VAL-LYS, PERIPLASMIC OLIGOPEPTIDE-BINDING PROTEIN, ... | | Authors: | Tame, J.R.H, Sleigh, S.H, Wilkinson, A.J. | | Deposit date: | 1999-07-14 | | Release date: | 1999-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and Calorimetric Analysis of Peptide Binding to Oppa Protein
J.Mol.Biol., 291, 1999
|
|
6ZM3
 
 | | The structure of an E2 ubiquitin-conjugating complex (UBC2-UEV1) essential for Leishmania amastigote differentiation | | Descriptor: | Putative ubiquitin-conjugating enzyme e2, Ubiquitin-conjugating enzyme-like protein | | Authors: | Burge, R.J, Dodson, E.J, Wilkinson, A.J, Mottram, J.C. | | Deposit date: | 2020-07-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Leishmania differentiation requires ubiquitin conjugation mediated by a UBC2-UEV1 E2 complex.
Plos Pathog., 16, 2020
|
|
2B18
 
 | | N-terminal GAF domain of transcriptional pleiotropic repressor CodY. | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor codY, ISOLEUCINE | | Authors: | Levdikov, V.M, Blagova, E, Joseph, P, Sonenshein, A.L, Wilkinson, A.J. | | Deposit date: | 2005-09-15 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of CodY, a GTP- and Isoleucine-responsive Regulator of Stationary Phase and Virulence in Gram-positive Bacteria.
J.Biol.Chem., 281, 2006
|
|
1H4Z
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1QMP
 
 | | Phosphorylated aspartate in the crystal structure of the sporulation response regulator, Spo0A | | Descriptor: | CALCIUM ION, Stage 0 sporulation protein A | | Authors: | Lewis, R.J, Brannigan, J.A, Muchova, K, Barak, I, Wilkinson, A.J. | | Deposit date: | 1999-10-04 | | Release date: | 1999-11-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylated aspartate in the structure of a response regulator protein.
J. Mol. Biol., 294, 1999
|
|
1H4X
 
 | | Structure of the Bacillus Cell Fate Determinant SpoIIAA in the Phosphorylated Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
5UCG
 
 | | Structure of the PP2C Phosphatase Domain and a Fragment of the Regulatory Domain of the Cell Fate Determinant SpoIIE from Bacillus Subtilis | | Descriptor: | Stage II sporulation protein E | | Authors: | Bradshaw, N, Levdikov, V, Zimanyi, C, Gaudet, R, Wilkinson, A, Losick, R. | | Deposit date: | 2016-12-22 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.906 Å) | | Cite: | A widespread family of serine/threonine protein phosphatases shares a common regulatory switch with proteasomal proteases.
Elife, 6, 2017
|
|
7ATR
 
 | |
2W40
 
 | | Crystal structure of Plasmodium falciparum glycerol kinase with bound glycerol | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GLYCEROL KINASE, ... | | Authors: | Schnick, C, Polley, S.D, Fivelman, Q.L, Ranford-Cartwright, L, Wilkinson, S.R, Brannigan, J.A, Wilkinson, A.J, Baker, D.A. | | Deposit date: | 2008-11-18 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure and Non-Essential Function of Glycerol Kinase in Plasmodium Falciparum Blood Stages.
Mol.Microbiol., 71, 2009
|
|
2W41
 
 | | Crystal structure of Plasmodium falciparum glycerol kinase with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL KINASE, ... | | Authors: | Schnick, C, Polley, S.D, Fivelman, Q.L, Ranford-Cartwright, L, Wilkinson, S.R, Brannigan, J.A, Wilkinson, A.J, Baker, D.A. | | Deposit date: | 2008-11-18 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure and Non-Essential Function of Glycerol Kinase in Plasmodium Falciparum Blood Stages.
Mol.Microbiol., 71, 2009
|
|
6TAA
 
 | | STRUCTURE AND MOLECULAR MODEL REFINEMENT OF ASPERGILLUS ORYZAE (TAKA) ALPHA-AMYLASE: AN APPLICATION OF THE SIMULATED-ANNEALING METHOD | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION | | Authors: | Swift, H.J, Brady, L, Derewenda, Z.S, Dodson, E.J, Turkenburg, J.P, Wilkinson, A.J. | | Deposit date: | 1992-08-21 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and molecular model refinement of Aspergillus oryzae (TAKA) alpha-amylase: an application of the simulated-annealing method.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
6I3G
 
 | | Crystal structure of a putative peptide binding protein AppA from Clostridium difficile | | Descriptor: | ABC transporter, substrate-binding protein, family 5, ... | | Authors: | Hughes, A.M, Wilkinson, A, Dodson, E. | | Deposit date: | 2018-11-06 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative peptide-binding protein AppA from Clostridium difficile.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3DD6
 
 | | Crystal structure of Rph, an exoribonuclease from Bacillus anthracis at 1.7 A resolution | | Descriptor: | Ribonuclease PH, SULFATE ION | | Authors: | Rawlings, A.E, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Wilson, K.S, Wilkinson, A.J, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2008-06-05 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | The structure of Rph, an exoribonuclease from Bacillus anthracis, at 1.7 A resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1JEU
 
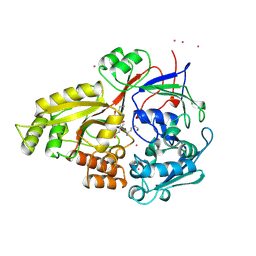 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KEK | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN, PEPTIDE LYS GLU LYS, URANYL (VI) ION | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1996-07-03 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The role of water in sequence-independent ligand binding by an oligopeptide transporter protein.
Nat.Struct.Biol., 3, 1996
|
|
1JEV
 
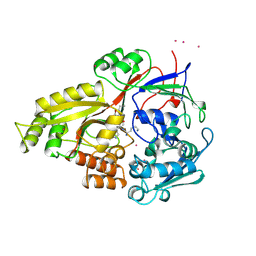 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KWK | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN, PEPTIDE LYS TRP LYS, URANYL (VI) ION | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1996-07-03 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The role of water in sequence-independent ligand binding by an oligopeptide transporter protein.
Nat.Struct.Biol., 3, 1996
|
|
1JET
 
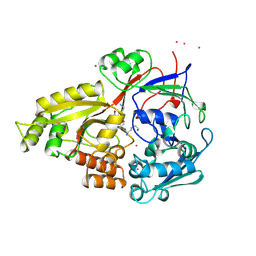 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KAK | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN, PEPTIDE LYS ALA LYS, URANYL (VI) ION | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1996-07-03 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The role of water in sequence-independent ligand binding by an oligopeptide transporter protein.
Nat.Struct.Biol., 3, 1996
|
|
3TGF
 
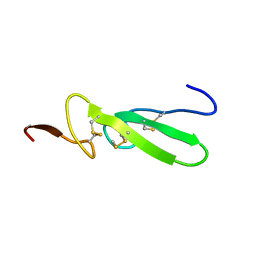 | | THE SOLUTION STRUCTURE OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA | | Descriptor: | TRANSFORMING GROWTH FACTOR-ALPHA | | Authors: | Harvey, T.S, Wilkinson, A.J, Tappin, M.J, Cooke, R.M, Campbell, I.D. | | Deposit date: | 1991-01-23 | | Release date: | 1993-04-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human transforming growth factor alpha.
Eur.J.Biochem., 198, 1991
|
|
3O9P
 
 | | The structure of the Escherichia coli murein tripeptide binding protein MppA | | Descriptor: | L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, Periplasmic murein peptide-binding protein, ZINC ION | | Authors: | Maqbool, A, Levdikov, V.M, Blagova, E.V, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2010-08-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Compensating Stereochemical Changes Allow Murein Tripeptide to Be Accommodated in a Conventional Peptide-binding Protein.
J.Biol.Chem., 286, 2011
|
|
