7GPU
 
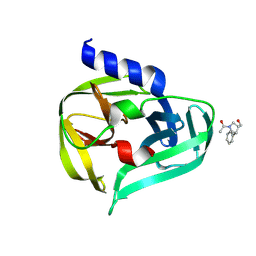 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with POB0122 | | Descriptor: | 1-[(3R,4R)-3-(hydroxymethyl)-4-phenylpyrrolidin-1-yl]ethan-1-one, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GQF
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z362020366 | | Descriptor: | DIMETHYL SULFOXIDE, Protease 3C, [1,2,4]triazolo[4,3-a]pyridin-3-amine | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GP0
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z385450668 | | Descriptor: | 2-(difluoromethoxy)benzene-1-sulfonamide, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GPM
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z979742720 | | Descriptor: | 5-(propan-2-yl)-3-[(2S)-pyrrolidin-2-yl]-1,2,4-oxadiazole, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GQ0
 
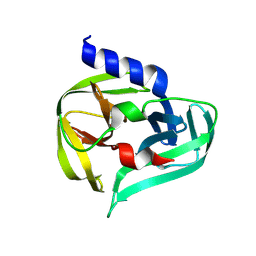 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with NCL-00025345 | | Descriptor: | 2-acetamido-N-(3-bromanylprop-2-ynyl)ethanamide, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GQB
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z44548882 | | Descriptor: | 1,1-diphenylmethanamine, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GQR
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z235343929 | | Descriptor: | 2-chloro-1,3-benzoxazole, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GPO
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with POB0066 | | Descriptor: | (3R,4S)-1-acetyl-4-phenylpyrrolidine-3-carboxylic acid, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GQP
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1198152494 | | Descriptor: | 1H-imidazole-5-carbonitrile, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
1XCI
 
 | | Mispair Aligned N3T-Butyl-N3T Interstrand Crosslink | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*(TTM)P*TP*TP*TP*CP*G)-3' | | Authors: | da Silva, M.W, Bierbryer, R.G, Wilds, C.J, Noronha, A.M, Colvin, O.M, Miller, P.S, Gamcsik, M.P. | | Deposit date: | 2004-09-02 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Intrastrand base-stacking buttresses widening of major groove in interstrand cross-linked B-DNA.
Bioorg.Med.Chem., 13, 2005
|
|
2MH6
 
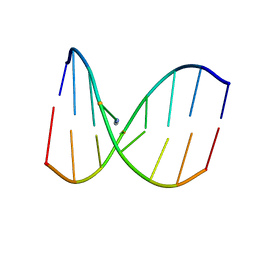 | | Solution structure of DNA duplex containing N3T-ethylene-N1I interstrand cross-link | | Descriptor: | 5'-D(*CP*AP*GP*TP*TP*CP*CP*A)-3', 5'-D(*TP*GP*GP*AP*IP*CP*TP*G)-3' | | Authors: | Denisov, A.Y, O'Flaherty, D.K, Noronha, A.M, Wilds, C.J. | | Deposit date: | 2013-11-18 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of an Ethylene Interstrand Cross-Linked DNA which Mimics the Lesion Formed by 1,3-Bis(2-chloroethyl)-1-nitrosourea.
Chemmedchem, 9, 2014
|
|
5TGG
 
 | |
4JRD
 
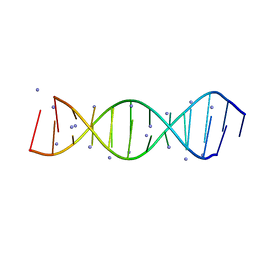 | | Crystal structure of the parallel double-stranded helix of poly(A) RNA | | Descriptor: | AMMONIUM ION, RNA (5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Safaee, N, Noronha, A.M, Kozlov, G, Rodionov, D, Wilds, C.J, Sheldrick, G.M, Gehring, K. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the parallel duplex of poly(A) RNA: evaluation of a 50 year-old prediction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3K54
 
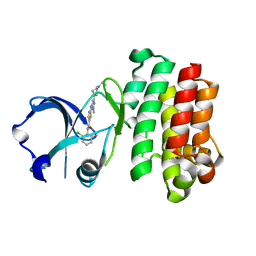 | | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggests a mechanism of activation for TEC family kinases. | | Descriptor: | N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Tyrosine-protein kinase BTK | | Authors: | Marcotte, D.J, Liu, Y.-T, Arduini, R.M, Hession, C.A, Miatkowski, K, Wildes, C.P, Cullen, P.F, Hopkins, B.T, Mertsching, E, Jenkins, T.J, Romanowski, M.J, Baker, D.P, Silvian, L.F. | | Deposit date: | 2009-10-06 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggest a mechanism of activation for TEC family kinases.
Protein Sci., 19, 2010
|
|
3GEN
 
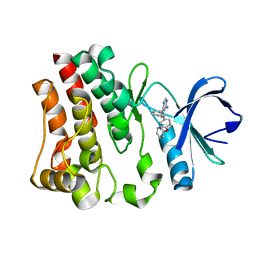 | | The 1.6 A crystal structure of human bruton's tyrosine kinase bound to a pyrrolopyrimidine-containing compound | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, Tyrosine-protein kinase BTK | | Authors: | Marcotte, D.J, Liu, Y.T, Arduini, R.M, Hession, C.A, Miatkowski, K, Wildes, C.P, Cullen, P.F, Hopkins, B, Mertsching, E, Jenkins, T.J, Romanowski, M.J, Baker, D.P, Silvian, L.F. | | Deposit date: | 2009-02-25 | | Release date: | 2010-01-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggest a mechanism of activation for TEC family kinases.
Protein Sci., 19, 2010
|
|
2LZW
 
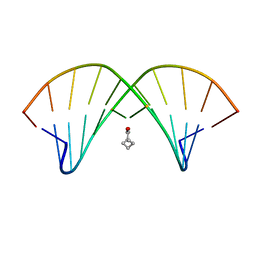 | | DNA duplex containing mispair-aligned O6G-heptylene-O6G interstrand cross-link | | Descriptor: | DNA (5'-D(*CP*GP*AP*AP*AP*GP*TP*TP*TP*CP*G)-3'), HEPTANE | | Authors: | Denisov, A.Y, McManus, F.P, Noronha, A.M, Wilds, C.J. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of interstrand cross-link repair by O 6 -alkylguanine DNA alkyltransferase.
Org.Biomol.Chem., 15, 2017
|
|
2LZV
 
 | | DNA duplex containing mispair-aligned O4U-heptylene-O4U interstrand cross-link | | Descriptor: | DNA (5'-D(*CP*GP*AP*AP*AP*UP*TP*TP*TP*CP*G)-3'), HEPTANE | | Authors: | Denisov, A.Y, McManus, F.P, Noronha, A.M, Wilds, C.J. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of interstrand cross-link repair by O 6 -alkylguanine DNA alkyltransferase.
Org.Biomol.Chem., 15, 2017
|
|
1I0T
 
 | | 0.6 A STRUCTURE OF Z-DNA CGCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', SPERMINE | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.6 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
2LSC
 
 | | Solution structure of 2'F-ANA and ANA self-complementary duplex | | Descriptor: | DNA (5'-D(*(CFL)P*(GFL)P*(CFL)P*(GFL)P*(A5O)P*(A5O)P*(UAR)P*(UAR)P*(CFL)P*(GFL)P*(CFL)P*(GFL))-3') | | Authors: | Martin-Pintado, N, Yahyaee, M, Campos, R, Noronha, A, Wilds, C, Damha, M, Gonzalez, C. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of double helical arabino nucleic acids (ANA and 2'F-ANA): effect of arabinoses in duplex-hairpin interconversion.
Nucleic Acids Res., 40, 2012
|
|
1PWF
 
 | | One Sugar Pucker Fits All: Pairing Versatility Despite Conformational Uniformity in TNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(2MU)P*(FA2)P*CP*GP*C)-3', SPERMINE | | Authors: | Pallan, P.S, Wilds, C.J, Wawrzak, Z, Krishnamurthy, R, Eschenmoser, A, Egli, M. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Why does TNA cross-pair more strongly with RNA than with DNA? an answer from X-ray analysis.
ANGEW.CHEM.INT.ED.ENGL., 42, 2003
|
|
1MA8
 
 | | A-DNA decamer GCGTA(UMS)ACGC with incorporated 2'-methylseleno-uridine | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*UMSP*AP*CP*GP*C)-3' | | Authors: | Teplova, M, Wilds, C.J, Wawrzak, Z, Tereshko, V, Du, Q, Carrasco, N, Huang, Z, Egli, M. | | Deposit date: | 2002-08-01 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Covalent incorporation of selenium into oligonucleotides for X-ray crystal structure determination via MAD: proof of principle
BIOCHIMIE, 84, 2002
|
|
1G2J
 
 | | RNA OCTAMER R(CCCP*GGGG) CONTAINING PHENYL RIBONUCLEOTIDE | | Descriptor: | 5'-R(*CP*CP*CP*(PYY)P*GP*GP*GP*G)-3', CALCIUM ION | | Authors: | Minasov, G, Matulic-Adamic, J, Wilds, C.J, Haeberli, P, Usman, N, Beigelman, L, Egli, M. | | Deposit date: | 2000-10-20 | | Release date: | 2000-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of an RNA duplex containing phenyl-ribonucleotides, hydrophobic isosteres of the natural pyrimidines.
RNA, 6, 2000
|
|
1I0G
 
 | | 1.45 A STRUCTURE OF THE A-DECAMER GCGTATACGC WITH A SINGLE 2'-O-FLUOROETHYL THYMINE IN PLACE OF T6, MEDIUM NA-SALT | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(125)P*AP*CP*GP*C)-3', SODIUM ION | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
1I0J
 
 | | 1.06 A STRUCTURE OF THE A-DECAMER GCGTATACGC WITH A SINGLE 2'-O-METHYL-3'-METHYLENEPHOSPHONATE (T23) THYMINE IN PLACE OF T6, HIGH CS-SALT | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(T23)P*AP*CP*GP*C)-3', CESIUM ION | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
1I0K
 
 | | 1.05 A STRUCTURE OF THE A-DECAMER GCGTATACGC WITH A SINGLE 2'-O-METHYL-[TRI(OXYETHYL)] THYMINE IN PLACE OF T6, MEDIUM CS-SALT | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(126)P*AP*CP*GP*C)-3', CESIUM ION | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
