1DM3
 
 | |
4ZDB
 
 | | Yeast enoyl-CoA isomerase (ScECI2) complexed with acetoacetyl-CoA | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, ACETOACETYL-COENZYME A, GLYCEROL, ... | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZDF
 
 | | Crystal structure of yeast enoyl-CoA isomerase helix-10 deletion (ScECI2-H10) mutant | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, GLYCEROL | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZDE
 
 | | Crystal structure of yeast D3,D2-enoyl-CoA isomerase F268A mutant | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, GLYCEROL, SULFATE ION | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZDD
 
 | | Structure of yeast D3,D2-enoyl-CoA isomerase bound to sulphate ion | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, SULFATE ION | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZDC
 
 | | Yeast enoyl-CoA isomerase complexed with octanoyl-CoA | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, GLYCEROL, OCTANOYL-COENZYME A, ... | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4B3J
 
 | |
4B3I
 
 | |
4BT9
 
 | |
4BT8
 
 | |
4BTA
 
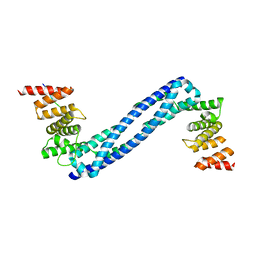 | | CRYSTAL STRUCTURE OF THE PEPTIDE(PRO-PRO-GLY)3 BOUND COMPLEX OF N- TERMINAL DOMAIN AND PEPTIDE SUBSTRATE BINDING DOMAIN OF PROLYL-4 HYDROXYLASE (RESIDUES 1-244) TYPE I FROM HUMAN | | Descriptor: | PROLINE RICH PEPTIDE, PROLYL 4-HYDROXYLASE SUBUNIT ALPHA-1 | | Authors: | Anantharajan, J, Koski, M.K, Pekkala, M, Wierenga, R.K. | | Deposit date: | 2013-06-14 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The Structural Motifs for Substrate Binding and Dimerization of the Alpha Subunit of Collagen Prolyl 4-Hydroxylase
Structure, 21, 2013
|
|
1MSS
 
 | |
4CQM
 
 | | Crystal structure of heterotetrameric human ketoacyl reductase complexed with NAD and NADP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CARBONYL REDUCTASE FAMILY MEMBER 4, ... | | Authors: | Venkatesan, R, SahTeli, S.K, Awoniyi, L.O, Jiang, G, Prus, P, Kastoniotis, A.J, Hiltunen, J.K, Wierenga, R.K, Chen, Z. | | Deposit date: | 2014-02-19 | | Release date: | 2014-09-10 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Insights Into Mitochondrial Fatty Acid Synthesis from the Structure of Heterotetrameric 3-Ketoacyl-Acp Reductase/3R-Hydroxyacyl-Coa Dehydrogenase.
Nat.Commun., 5, 2014
|
|
4BTB
 
 | |
4CQL
 
 | | Crystal structure of heterotetrameric human ketoacyl reductase complexed with NAD | | Descriptor: | CARBONYL REDUCTASE FAMILY MEMBER 4, ESTRADIOL 17-BETA-DEHYDROGENASE 8, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Venkatesan, R, Sah-Teli, S.K, Awoniyi, L.O, Jiang, G, Prus, P, Kastaniotis, A.J, Hiltunen, J.K, Wierenga, R.K, Chen, Z. | | Deposit date: | 2014-02-19 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Insights Into Mitochondrial Fatty Acid Synthesis from the Structure of Heterotetrameric 3-Ketoacyl-Acp Reductase/3R-Hydroxyacyl-Coa Dehydrogenase.
Nat.Commun., 5, 2014
|
|
8OPX
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Trehalose (Fragment-B-TRE) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, Putative acyltransferase Rv0859, SULFATE ION, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQO
 
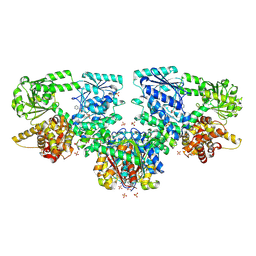 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-49 | | Descriptor: | GLYCEROL, Probable fatty oxidation protein FadB, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQT
 
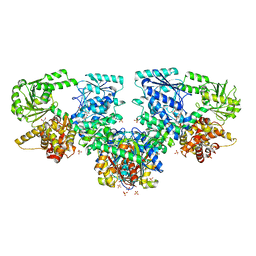 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-91 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-bromanylbenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OPU
 
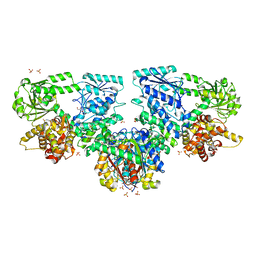 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Sulfamethoxazole (Fragment-B-E1) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OPW
 
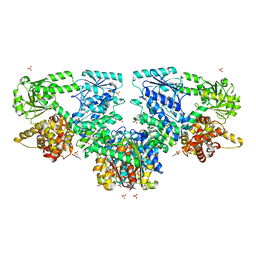 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Caffeine (Fragment-B-51) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, CAFFEINE, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OPY
 
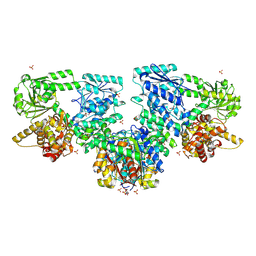 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-B-DNQ | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 6,7-DINITROQUINOXALINE-2,3-DIONE, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQS
 
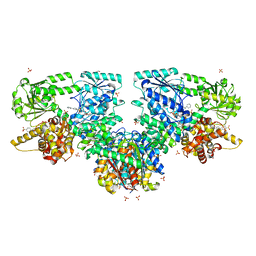 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-83 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-phenylbenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQV
 
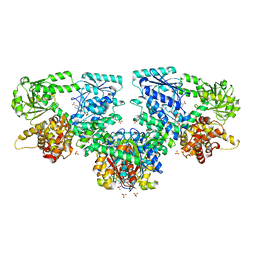 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-109 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-nitrobenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQL
 
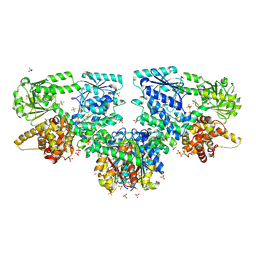 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-1 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Hexafluorophosphate anion, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQQ
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-79 | | Descriptor: | 2-fluoranyl-5-sulfo-benzoic acid, 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
